Chung-Chau Hon
@chungchau_hon
Team Leader @RIKEN_IMS Japan, working on stuffs somewhere between DNAs and RNAs in mostly single-cells, about disease genetics and gene regulation.
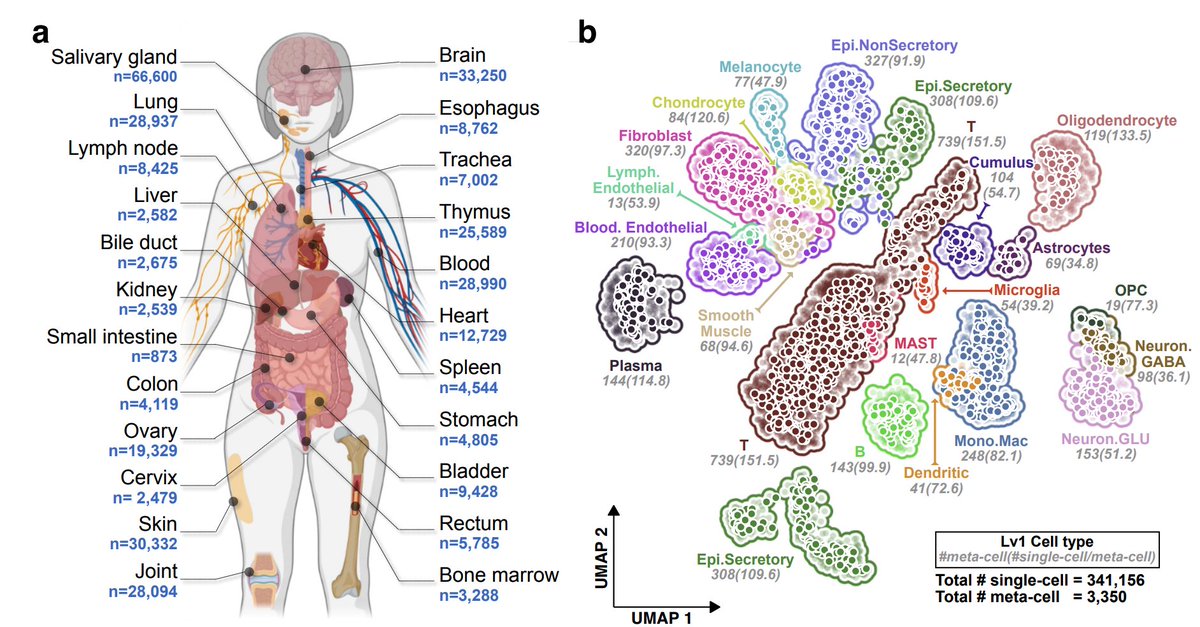
1/ Excited to share our latest work on mapping the transcribed cis-regulatory elements (tCREs) in single-cells across 23 human tissues for dissecting trait heritability with cellular contexts. doi.org/10.1101/2023.1…
🚀 Our perspective is out in @Nature! We present a roadmap for Multimodal Foundation Models (MFMs) — large AI models pretrained across multi-omics and multi-timepoint data — to serve as the computational backbone for building virtual cells. Read the full paper in Nature:…
Excited to share our new preprint on "CFC-seq & enhancer-derived transcription". We have identified ~24,000 eRNAs and their transcription are correlated with enhancer structures. Read more on @biorxivpreprint: [doi.org/10.1101/2024.1…]
Last talk. For today, @chungchau_hon talking about the cell atlas cantered on regulatory elements detected by 5end sequencing modified from biorxiv.org/content/10.110… Clearly 5’ rna sequencing advantageous to detect regulatory elements. #HGM24
Automating UMAP parameter tuning nature.com/articles/s4146…

Excellent postdocs positions to work at RIKEN. About 70 positions. Salaries up. Call for applications for the position of Special Postdoctoral Researcher (SPDR) for FY 2025 | RIKEN riken.jp/en/careers/pro…
I'm looking for an intern! 🐣 If you just finished your master or are about to and want to do a 6 months+ project working with #organoids, #optogenetics, #singlecell and #spatial omics, DM or drop an email in the next couple of weeks
Bonkers paper! From TF cooperatively and interdependence to crystal structure to biochemistry to human genetics! Tour de force from Joanna Wysocka's team including @seungsookim10, @jussitai, @snaqvi1990, and others: cell.com/cell/fulltext/…
Here are seven technologies we think we all need to watch in 2024 @nature nature.com/articles/d4158…
An absolutely amazing book
Why do we get certain diseases whereas others do not exist? This new book builds a foundation for systems medicine. Starting from basic laws, it derives why hormone, immune and aging circuits are built the way the are, culminating in a periodic table of diseases.
Last couple of days to apply for this postdoc position to generate deep learning models and expand the great JASPAR and UniBind databases @NCMMnews! It's a fantastic project in a very supportive group with outstanding collaborators!
🧬Opening for a postdoc in my group @NCMMnews @UniOslo @UniOslo_MED @unioslo_mn to generate deep learning models to enhance JASPAR @jaspar_db & UniBind, boosting gene regulation research. Project in collaboration with @anshulkundaje, @WyWyWa. Plz share jobbnorge.no/en/available-j…
Proud to have lead this work alongside Nathan! This paper represents a huge effort between the Ren lab, @JoeEcker lab, @CEpigenomics, and many more! nature.com/articles/s4158…
Dividing out quantification uncertainty allows efficient assessment of differential transcript expression with edgeR. #DifferentialExpression #RNAseq #TranscripLevelQuantification @nar_open academic.oup.com/nar/advance-ar…
Important benchmarking effort! Let the data talk. Too much hype, litigation and marketing in this space. Great to see. Thank you to the @GoodsBrittany and Farhi teams for this important preprint.
Congrats Sami and team! Systematic benchmarking of imaging spatial transcriptomics platforms in FFPE tissues biorxiv.org/content/10.110…
Decoding human genetic variation using a synthetic paradigm go.nature.com/3uNVT28 #JournalClub by @Aashiq_Kachroo
Identifying cell states in single-cell RNA-seq data at statistically maximal resolution buff.ly/47PTZNg Anyone tried this? I heard that it takes a long time to run...
Single cell transcriptome of 17 immune cell types x 86 cytokines in mouse lymph nodes in vivo.
✨#ImmuneDictionary✨is out today in @Nature! Paper nature.com/articles/s4158… Software immune-dictionary.org We created scRNA-seq dictionary of 17+ immune cell types responding to 86 cytokines in vivo, discovered the immune system is far more complex than previously known 1/
PROUD to PRESENT⭐️⭐️⭐️! Our latest work @humancellatlas generating an Atlas of Inflammation across acute/chronic inflammatory diseases. Interpretable ML for Biomarker selection and classification. #diagnostics CELLS in CIRCULATION- a powerful resource for #precisionmedicine
THRILLED to share “An Interpretable Inflammation Landscape of Circulating Immune Cells” from @hoheyn lab @cnag_eu . ~2M PBMC🩸, 356👤, 18 Diseases ML for Biomarker Discovery & Patient Classification doi.org/10.1101/2023.1… Inflammation Atlas for @humancellatlas #tweetorial (1/n)
Happy to share that our methodology to assay thousands of regulatory regions and/or genetic variants with Massively parallel reporter assays (MPRAs) is out: tinyurl.com/3r7tyz5p @STARProtocols Thanks to Kate, Chiara, and John!
THRILLED to share “An Interpretable Inflammation Landscape of Circulating Immune Cells” from @hoheyn lab @cnag_eu . ~2M PBMC🩸, 356👤, 18 Diseases ML for Biomarker Discovery & Patient Classification doi.org/10.1101/2023.1… Inflammation Atlas for @humancellatlas #tweetorial (1/n)