
Javier Santoyo
@jsantoyo
Genomics & Bioinformatics Scientist at @EdinburghUni. Head of @edgenome | #Omics #NGS #LongReads | Views my own | Also @ http://jsantoyo.bsky.social
MKDESIGNER and TASEQ: a set of tools for plant genotyping by targeted amplicon sequencing. #PlantGenotyping #TargetedAmpliconsSequencing #Bioinformatics #BMCbioinformatics bmcbioinformatics.biomedcentral.com/articles/10.11…

PhyloScape: interactive and scalable visualization platform for phylogenetic trees. r#PhylogeneticTees #Visualization #Bioinformatics #BMCbioinformatics bmcbioinformatics.biomedcentral.com/articles/10.11…

clonevdjseq: A workflow and bioinformatics management system for sequencing, archiving, and analysis of VDJ sequences from clonal libraries. #VDJ #Sequencing #Bioinformatics #BMCbioinformatics bmcbioinformatics.biomedcentral.com/articles/10.11…

Evaluating the representational power of pre-trained DNA language models for regulatory genomics.#gLM #Genomics #Bioinformatics @GenomeBiology genomebiology.biomedcentral.com/articles/10.11…

Advancing metagenomic classification with NABAS+: a novel alignment-based approach. #Metagenomics #Genomics #Bioinformatics #NARgenomicsAndBioinformatics academic.oup.com/nargab/article…

Comparison of single-cell long-read and short-read transcriptome sequencing via cDNA molecule matching: quality evaluation of the MAS-ISO-seq approach. #SingleCell #scRNAseq #ShortRead #LongRead #SequencingComparison #NARgenomicsAndBioinformatics academic.oup.com/nargab/article…

Chevreul: an R bioconductor package for exploratory analysis of full-length single cell sequencing. #SingleCell #scRNAseq #FullLengthRNA #GigaByte gigabytejournal.com/articles/158

Integrating comparative genomics and risk classification by assessing virulence, antimicrobial resistance, and plasmid spread in microbial communities with gSpreadComp. #Genomics #MicrobialGenomics #AMR @GigaScience academic.oup.com/gigascience/ar…

eNRSA: a faster and more powerful approach for nascent transcriptome analysis. #Transcriptomics #NascentTranscripts #Genomics #Bioinformatics @GigaScience academic.oup.com/gigascience/ar…

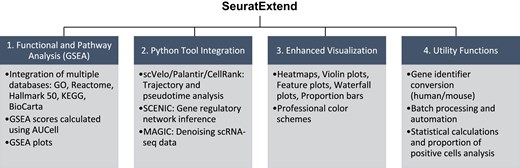
SeuratExtend: streamlining single-cell RNA-seq analysis through an integrated and intuitive framework. #SingleCell #scRNAseq #Genomics #Bioinformatics @GigaScience academic.oup.com/gigascience/ar…
Tsbrowse: an interactive browser for Ancestral Recombination Graphs. #AncestralRecombinationGraphs #Genomics #Bioinformatics academic.oup.com/bioinformatics…

Identifying DNA methylation types and methylated base positions from bacteria using nanopore sequencing with multi-scale neural network.#DNAmethylation #BacterialDNA #Nanopore #Sequencing #Genomics #Bioinformatics @nanopore academic.oup.com/bioinformatics…

BugBuster: A novel automatic and reproducible workflow for metagenomic data analysis. #Metagenomics #DataAnalysis #AutomaticPipeline #Genomics #Bioinformatics @BioinfoAdv academic.oup.com/bioinformatics…

Genome Evaluation Pipeline (GEP): A fully-automated quality control tool for parallel evaluation of genome assemblies. #GenomeAssembly #QualityControl #Snakemake #Genomics #Bioinformatics @BioinfoAdv academic.oup.com/bioinformatics…

PangenePro: an automated pipeline for rapid identification and classification of gene family members. #Pangenomes #GeneClassification #Genomics #Bioinformatics @BioinfoAdv academic.oup.com/bioinformatics…

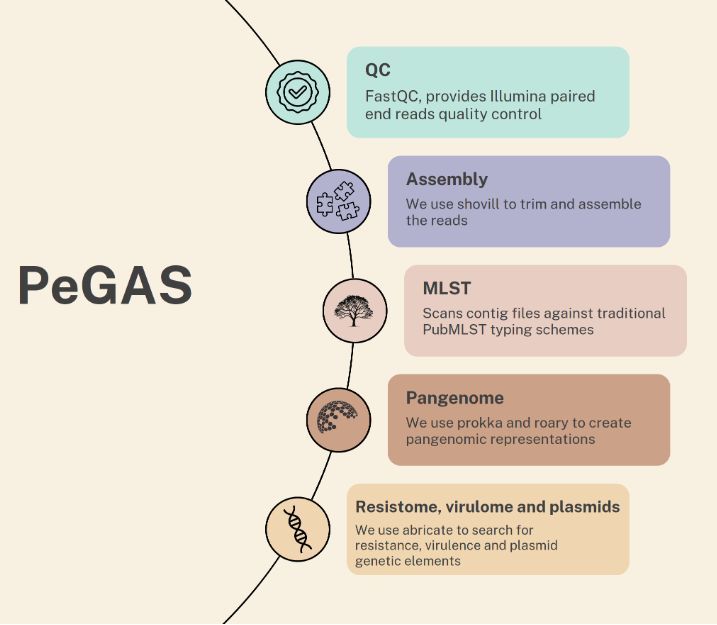
PeGAS: A Versatile Bioinformatics Pipeline for AMR, Virulence and Pangenome Analysis. #AMR #PangenomeAnalysis #Genomics #Bioinformatics @BioinfoAdv academic.oup.com/bioinformatics… Github: github.com/liviurotiul/Pe…
Benchmarking Peak Calling Methods for CUT&RUN. #CUTandRUN #Benchmarking #Genomics #Bioinformatics academic.oup.com/bioinformatics…

CCRR: A User-Friendly Platform for Analyzing Complex Chromosomal Rearrangements in Tumors. #SVs #CNV #ChromosomalRearrangements #Genomics #Bioinformatics academic.oup.com/bioinformatics…

Agptools: a utility suite for editing genome assemblies. #GenomeAssembly #AssemblyEditing #AGPfiles #Genomics #Bioinformatics academic.oup.com/bioinformatics…

Using >130 human & 12 primate haplotypes, we reconstruct the chromosome 22q11.2 evolution to identify haplotype structures linked to deletions or inversions, explaining the lower prevalence of 22q11.2 deletion syndrome in individuals of African descent. biorxiv.org/content/10.110…