Adam Green
@adamlewisgreen
building interpretable biological simulators
Despite staggering amounts of data, when it comes to understanding biology, we still see through a glass, darkly. As a first step towards elucidation, we at Markov built a virtual cell and tools to peer inside, probe, and perturb its mind, allowing us to better understand it.
yapping continues to be an underrated career strategy (the occasional reference or two helps as well)


it's only a causal model if it comes from the Pearlian region and receives the "model identification guaranteed" appellation, otherwise it's just sparkling zero-shot out-of-distribution generalization
A model trained with a frequentist objective using i.i.d. observational train and validation data is almost surely converging to the non-causal model of the underlying data generating process. That is because using short cuts/spurious correlations leads to a lower loss than not,…
I’d love to hear biologists weigh in on the fundamental question: can you predict the impact of cell perturbations better by studying the natural variation in a population of healthy cells (more data), or by studying cells that have been perturbed genetically or chemically?
virtual cells are currently bottlenecked by compute, not novel data: drug discovery is an iterative search process (design, test, analyze) through therapeutic design space guided by a dynamics model directly trawling this therapeutic space with large hypothesis-free…
learning to predict the next token teaches your model a lot more than just fancy n-gram statistics, it turns out
To me, this is a wildly unexpected take. So it's especially interesting that Markov Bio is currently (July 19 12pm) atop the Arc competition leaderboard. virtualcellchallenge.org/leaderboard
the brown m&m test of virtual cell papers: do you explicitly compute loss on observed zeros
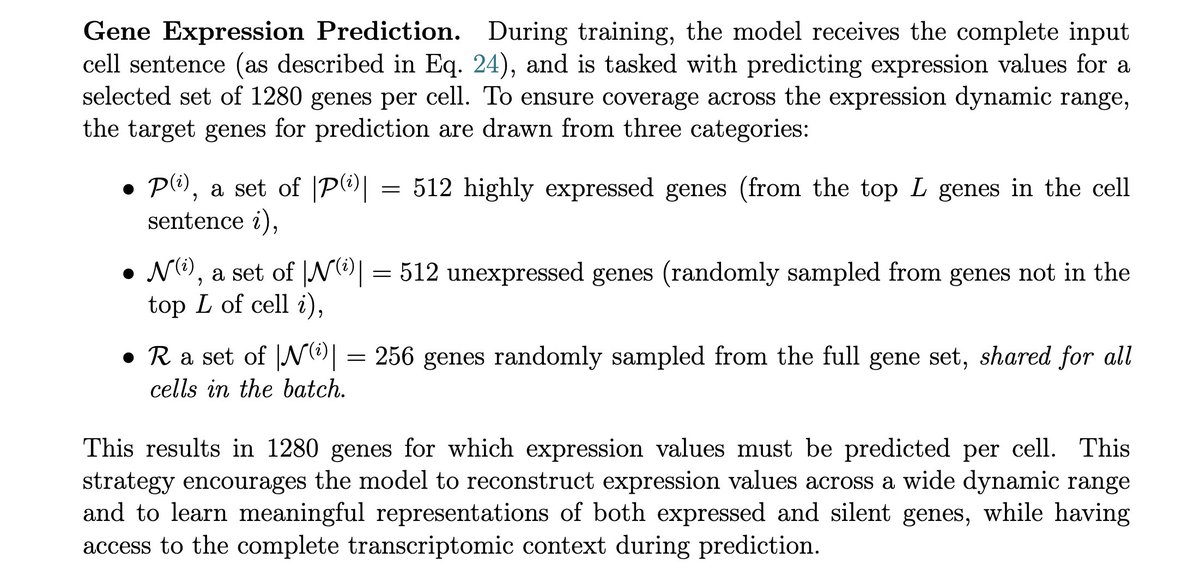
building a useful virtual cell is a scale and skill (and vision) issue. others have still not yet internalized this and it shows. exciting open-source developments on this front coming from Markov in the next few weeks
virtual cells are currently bottlenecked by compute, not novel data: drug discovery is an iterative search process (design, test, analyze) through therapeutic design space guided by a dynamics model directly trawling this therapeutic space with large hypothesis-free…
We're bringing on a guest who I've wanted to have on for a long time — the one and only @owl_posting, joining me and @AndrewE_Dunn. See you here at noon! youtube.com/live/S2AQ1mFhT…
if only there were a system that could generate truly novel therapeutic hypotheses not found in the literature, elucidate their mechanism, and use inference-time compute to iteratively optimize candidate molecules to cause any desired delta in biological state...
Today, we’re announcing the first major discovery made by our AI Scientist with the lab in the loop: a promising new treatment for dry AMD, a major cause of blindness. Our agents generated the hypotheses, designed the experiments, analyzed the data, iterated, even made figures…
We are excited to have @adamlewisgreen to join us next Tuesday. Adam is the founder at Markov Bioscience and he has deep insights into virtual cell models, mechanistic interpretability and so on. Join us next Tuesday 2:30PM at Gates 415!
just one more striped hyena block bro, I promise. DNA is all you need dog, trust me. the only thing we're missing is ring attention, I swear. once we have the context length to capture long-range interactions we'll solve mammalian genome modeling. yeah yeah I know the human…
The major way through which SVs affected gene expression was through copy number changes. Reshuffling the relative location of genes through inversions or translocations did not influence their expression. Expression of genes nearby the SVs was rarely affected.
tfw you actually simulate 500 million years of evolution in silico by teaching your pretrained virtual cell to self-organize into stable multicellular systems using RL against the ultimate verifiable reward—survival—thereby regularizing it to better match the evolutionarily…
We are super excited to share our seminar schedule for the next few months - join us at Stanford campus to learn about the latest in AI + Biomedicine! Anyone is welcome! snap.stanford.edu/ai-bio-seminar/
virtual cells are currently bottlenecked by compute, not novel data: drug discovery is an iterative search process (design, test, analyze) through therapeutic design space guided by a dynamics model directly trawling this therapeutic space with large hypothesis-free…