Taka Inoue
@TakaInoue5
Associate Professor, Kyoto University
@RyuichiroNakato, @TakaInoue5 and I are organizing a session at MBSJ2024 on "Large-scale computational epigenomics for the elucidation of genomic regulatory mechanisms." Please consider submitting an abstract, deadline July 31! aeplan.jp/mbsj2024/en-sy…
🚨 Job opportunity, please RT! 🚨 My lab in Kyoto 🇯🇵 is looking to recruit 2 postdocs to work on comparative epigenomics, transposable elements, and/or related topics. Come work with @TakaInoue5 and me @Ashbi_KyotoU ! ashbi.kyoto-u.ac.jp/wp/wp-content/…
Happy to share our paper is published in NAR today !!! @NadavAhituv @YosefLab @TalAshuach @TakaInoue5 @RutgersCABM academic.oup.com/nar/advance-ar…
Congrats to @FengYuanqing @Xieningjing0612 and many collaborators for publication of their study integrating GWAS, selections scans in the San, and functional genomics to identify regulatory variants and target genes influencing skin color in Africans! nature.com/articles/s4158…
【拡散希望】UCSF @NadavAhituv 研究室では、遺伝学・ゲノミクス研究に従事していただける博士研究員の方を募集しています。マウスを用いた実験経験のある方を特に探しています。Nadav はサポーティブなPI で、大変充実した研究生活を送れています。Jrec-inリンク先👇 jrecin.jst.go.jp/seek/SeekJorDe…
We all take repeat annotation in the human genome for granted but are we sure it's correct? In a 🚨new paper 🚨 from the lab, @xunchen85 shows that up to 30% of primate ERVs/LTRs instances are mis-annotated.
Super excited to share our latest #preprint on the TE classification/annotation and regulatory evolution from our lab @guilbourque at @Ashbi_KyotoU. Check it out here! 🧵 Cryptic endogenous retrovirus subfamilies in the primate lineage doi.org/10.1101/2023.1…
Our preprint is online!! biorxiv.org/content/10.110… @NadavAhituv @NirYosef4 @TakaInoue5 @TalAshuach
Interested to hear about the human pangenome? I'll be at the CHU de Quebec - Université Laval next week!
Interested in discovering the genetic changes that made us human? Our lab is looking for students and postdocs! 👩🔬👨🔬 gokhmanlab.com please RT!
The question of what makes us human remains enigmatic, but we investigate how enhancer hijacking, or fondly, "HARjacking", may have played a role in human-specific genome evolution in our study now out in @ScienceMagazine ! science.org/doi/10.1126/sc…
MPRA on 680,000 sequences comprehensively covering all active regulatory elements in 3 cell lines (HepG2,K562,iPSCs). Tour de force by @vagar112 & @TakaInoue5, @JShendure and @KircherLab as part of our @ENCODE_NIH project. biorxiv.org/content/10.110…
A really beautiful review. MPRAs are an incredibly powerful tool and it's time to make the most of them in evo studies!
「iPS細胞でヒトの進化を再現したい」 CiRA ニュースレターVol.52のCOMMUNITY(OB・OG紹介)では、かつて山中伸弥研究室に所属していた京都大学ASHBi 博士研究員の大貫茉里さんに、これまでのキャリアと育児をしながら行ったドイツでの研究についてお話を伺いました。 cira.kyoto-u.ac.jp/j/pressrelease…
Machine learning dissection of human accelerated regions in primate neurodevelopment cell.com/neuron/fulltex…
The hypothalamus is amazing! Here, we dissect it to single-cell parts link them to function and show how some of them can lead to obesity. Amazing work by Hai Nguyen, Candace Chan and collaboration with Christian Vaisse’s lab. biorxiv.org/content/10.110…
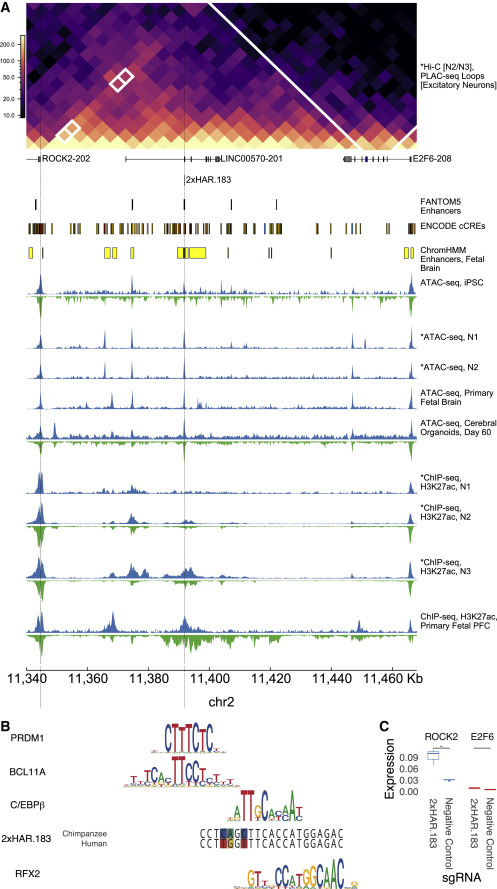
Three-dimensional genome re-wiring in loci with Human Accelerated Regions biorxiv.org/cgi/content/sh… #biorxiv_evobio
Exciting to share our latest works on #transposons #epigenomics and #viruses with an amazing group of scientists lead by @guilbourque, @LB_Barreiro, and Tomi Pastinen. tinyurl.com/4mp6exhu
Come visit the Inoue lab website in ASHBi Bourque group😀 ashbi.kyoto-u.ac.jp/lab-sites/inou… Japanese ver. ashbi.kyoto-u.ac.jp/lab-sites/inou…