
Marc Haber مارك
@MarcHaber
Associate professor in population genetics @unibirmingham
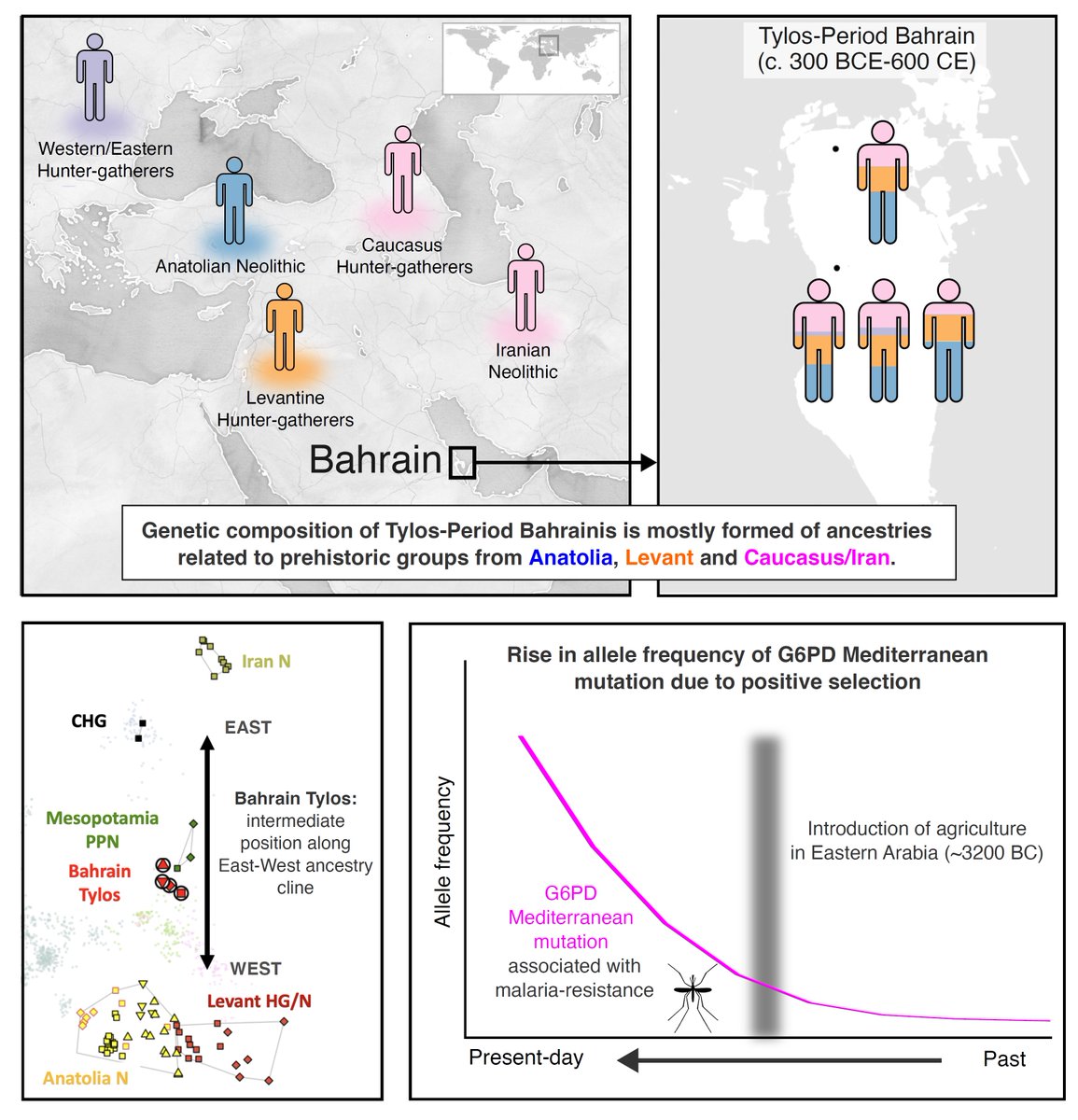
We publish today the first ancient whole-genome sequences from Arabia and show how they inform on ancestry and disease in the Middle East 🧬☠️ #aDNA #MiddleEast #Bahrain cell.com/cell-genomics/…
Analysis of fats and oils on pottery has suggested that Bronze Age communities in eastern Arabia consumed more plants than indicated by the archaeological record. thenationalnews.com/news/uae/2025/…
Thrilled to announce that our book chapter was published by @routledgebooks in #OpenAccess. This chapter is part of an outstanding 740-page book that examines the early events of the Bantu expansion in Central Africa from multidisciplinary perspectives. 🧵taylorfrancis.com/chapters/oa-ed…
Postdoc position opening in my group! Research projects: pangenomes for diverse organisms, genome evolution, biocomputing, language models. Please reach out if interested!
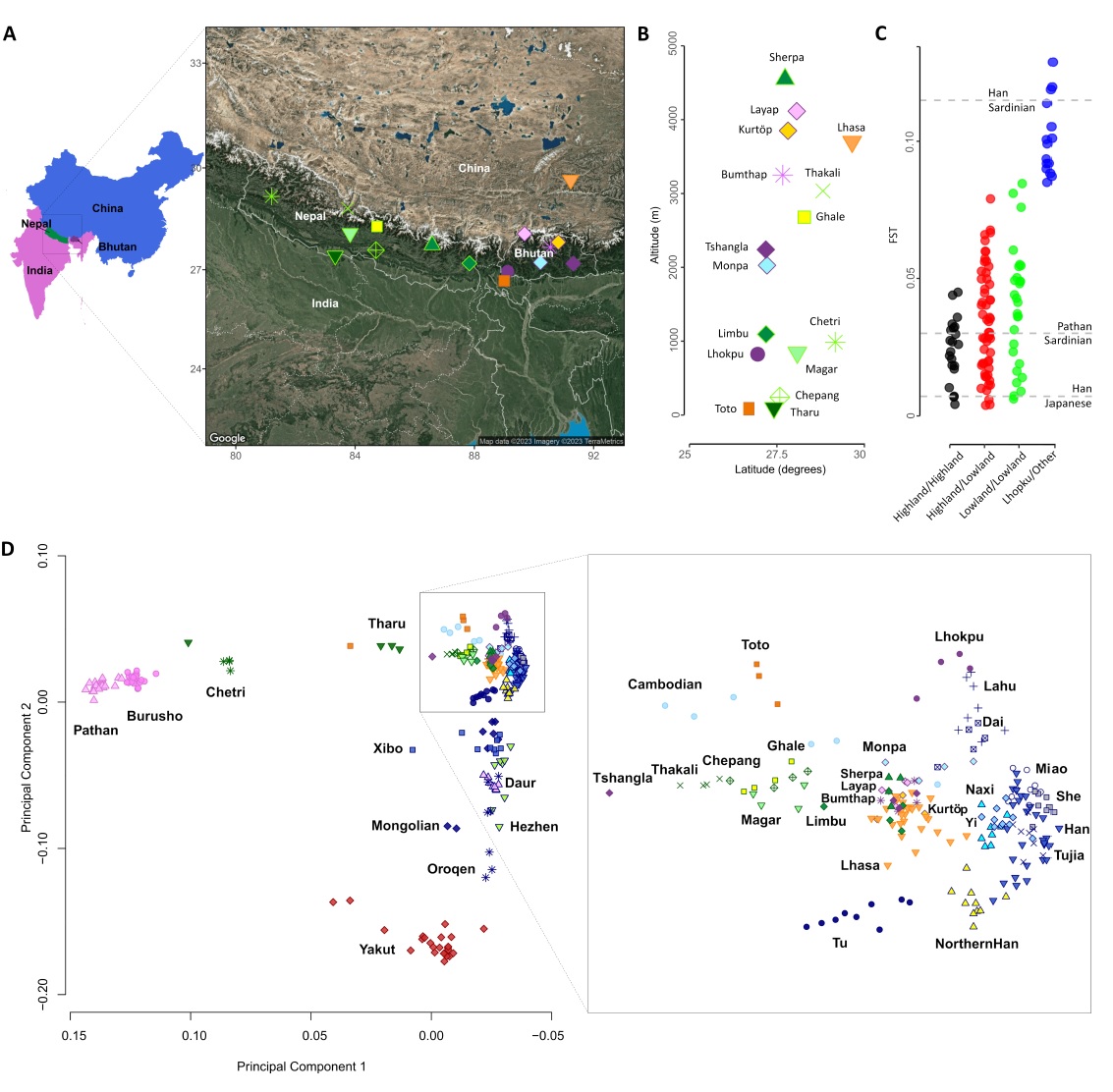
Our genomics experts have worked with partners from Europe, Asia, and the Middle East to uncover how human populations adapted, survived, and diversified in the Himalayas - one of the most extreme and challenging environments on Earth birmingham.ac.uk/news/2025/geno…
Our new paper on genetic diversity in the Himalayas, when it began and how it may have supported survival in extreme environments sciencedirect.com/science/articl…

Our new paper on ancient DNA to address the spread of Uralic languages now out in @nature (we started working on this in 2018). Many interesting results, including a sample from ~16k years ago Yakutia that can be modeled as an unadmixed Native American. nature.com/articles/s4158…
A 4-year postdoc position in population genomics is available in my group at the University of East Anglia, to work on a project sequencing a thousand fox genomes across rural and urban environments in the UK. vacancies.uea.ac.uk/vacancies/1593…
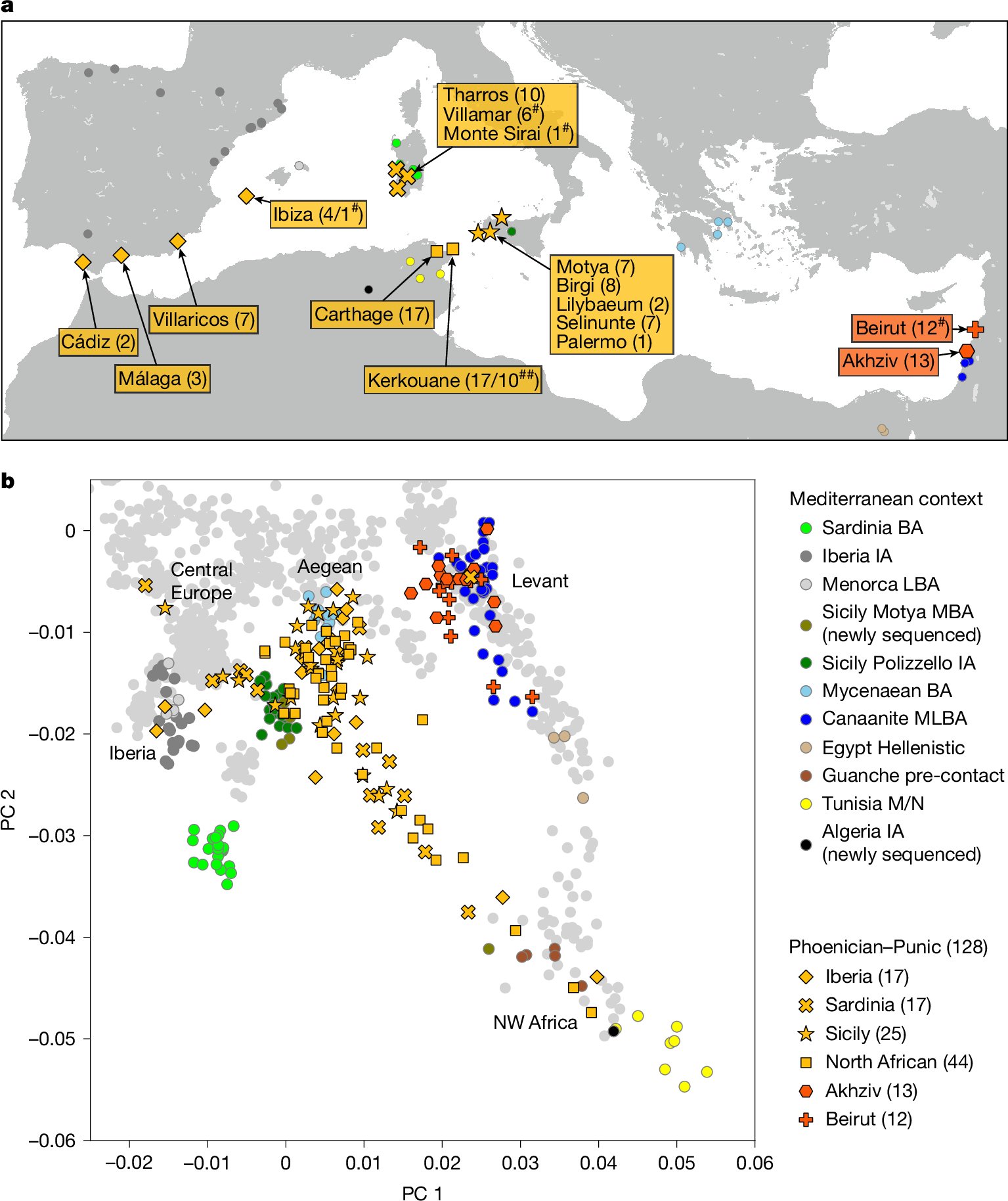
"We added previously published sequences for 20 indivdiuals [sic] confidently classified as Phoenician or Punic (7 from 2 Sardinian sites, 1 from Ibiza and 12 from Beirut)" But 2 of those in Beirut were an Egyptian mom and her admixed son nature.com/articles/s4158…
Punic people were genetically diverse with almost no Levantine ancestors nature.com/articles/s4158…
How the Emirati Genome Project could lead to advances in personalised medicine. Thanks to @MarcHaber of @birminghamdubai and Prof Habiba Al Safar of @KhalifaUni for the comments. thenationalnews.com/news/uae/2025/…
Our perspective on pangenomes in diverse populations and their critical role in advancing Precision Medicine nature.com/articles/s4159…
Our papers, out today in @nature, show how ancient DNA from the Eneolithic and Bronze Age steppe points to a North Pontic origin of the Indo-European language family and a Caucasus-Lower Volga (CLV) origin of Indo-Anatolian (inclusive of the now extinct Anatolian languages). 1/
Ancient genomics support deep divergence between Eastern and Western Mediterranean Indo-European languages biorxiv.org/content/10.110…
We're looking for two people to join our Research Data Science Service in the @uob_bear team at @unibirmingham, funded by our Institute for Data and AI. Come and work on innovative research projects all across our world-class university: blog.bham.ac.uk/bear/2024/11/2…
We sequenced Himalayan populations to understand how they formed and adapted to their unique environment. Our findings are now posted on bioRxiv for comments and suggestions 🧬🏔️ #Genetics #Himalayas #Tibet biorxiv.org/content/10.110…
Demographic history and genetic variation of the Armenian population: The American Journal of Human Genetics cell.com/ajhg/fulltext/…

In a new preprint led by @TheNikhilMilind, we explored a fascinating paradox: For many traits the number of duplications or loss-of-function (LoF) mutations is correlated with phenotype. Curiously, for most traits, the AVERAGE direction of LoFs and Dups is the SAME. Why?
Our new paper on the dynamic ancient genomic history of the aurochs is out in Nature. Well done to all, esp. Conor Rossi. Free download link. rdcu.be/dYAV3
Deadline 5 November!
2025 PhD-student opportunity in our lab (skoglundlab.org) open! This year I am particularly looking for candidates interested in computational ancient human genomics: genetic history and selection. crick.ac.uk/careers-study/… Those with an exceptional interest in other…
Our study on ancient Anatolian sheep 🐑 genomes 🧬 is finally out!!! Tons of insights into the early domestication processes in Anatolia and some interesting bits to look into with further research across Eurasia. Check the paper by Damla Kaptan et al: doi.org/10.1093/molbev…