Alexandre Marand
@Marand_Lab
Assistant Professor @UMich. Chromatin biology, cis-regulatory dynamics, plant genomes.
Mapping genetic modifiers of epimutation rates reveals a punctuated-equilibrium model of CG methylome evolution biorxiv.org/content/10.110…
Our paper is now in @NatureBiotech! Topological velocity inference from spatial transcriptomic data. TopoVelo infers the direction of differentiation/migration; quantifies spatial cell influence; and identifies morphology changes during differentiation. 🧵rdcu.be/ewqMB
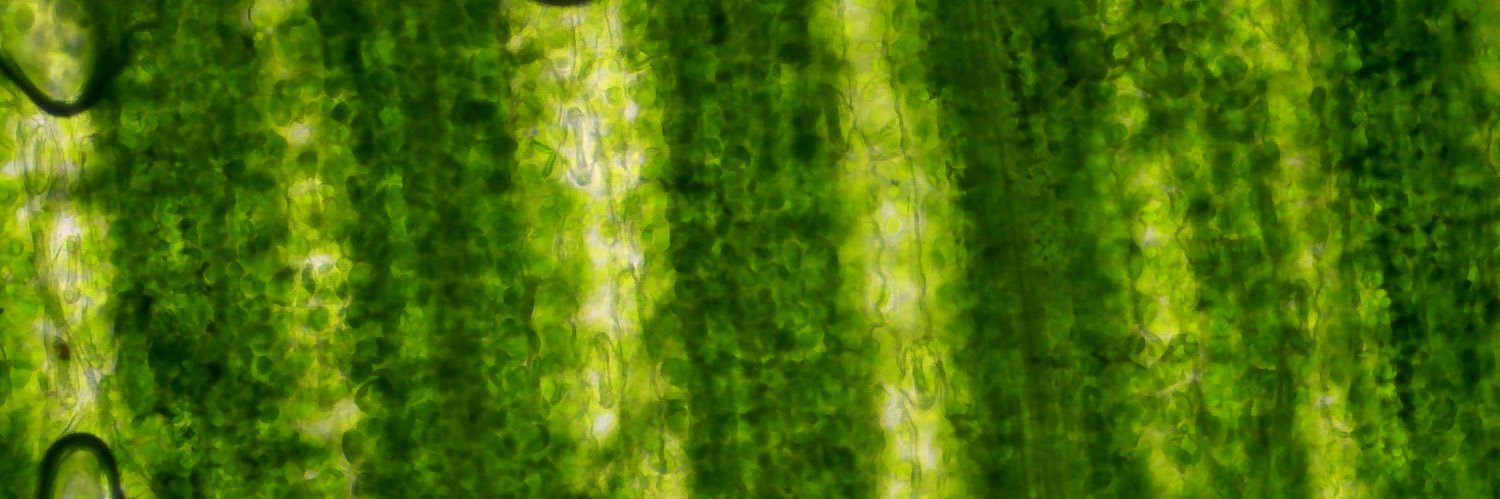
We're hiring! Need a microscopy wizard to help build the world’s first synthetic plant chromosome! Bring your lights, lasers, & lens-craft to characterise new artificial chromosomes. Join our ARIA-funded project at @uwanews in Perth & @plants4space. Apply: bit.ly/3GKSRSA
In the latest issue! LoxCode in vivo barcoding reveals epiblast clonal fate bias to fetal organs dlvr.it/TLtbl6
scMGCL: accurate and efficient integration representation of single-cell multiomics data academic.oup.com/bioinformatics…
🚨 New preprint alert! 🚨 We used single-cell RNA-seq + GRN analysis to uncover novel morphogenic genes that boost #maize transformation frequency 🌽 biorxiv.org/content/10.110… Thanks to @plaza_genomics and all other co-authors! #PlantScience #scRNAseq
I'll be joining the Department of Plant Biology at Michigan State University in Mid August as an Assistant Professor. cxli233.github.io/cxLi_lab/
Meet The Speakers of the Single-Cell Approaches in Plant Biology GRC Conference! To Apply to the GRC: lnkd.in/gvkMxuKR To Learn More About Dr. Nobori's Research: tatsuyanobori.com
New data from the Bob Schmitz lab (Marand 2025) on cell type-specific cis-regulation in maize! This study, in @ScienceMagazine, reveals how DNA changes impact expression and maize traits. Explore the 176 tracks on our browser (Epigenetics and DNA Binding) science.org/doi/10.1126/sc…
Defining “what is good” is central to both AI and Science. Despite massive efforts towards building the Virtual Cell, our recent @NatureBiotech article reflects on whether current gold standards (i.e., evaluation metrics) still hold up. nature.com/articles/s4158…
What makes a good cell embedding🧬? Is a higher score always better? Not quite! In this new preprint with @Jure & #Aviv, we ... [1/3] biorxiv.org/content/10.110…
We're excited to share PlantCAD browser tracks: a new, unpublished dataset integrating AI techniques for plant genome analysis. Explore it here: github.com/andorfc/PlantC… We welcome the community's feedback and innovative use cases for this data!
Development of this new genetic tool was sparked by a spontaneous discussion at a scientific conference. One two-year NSF EAGER grant to jump-start the work and 5 more years of "side projects" by 18 talented scientists made EBSn possible. biorxiv.org/content/10.110…
Wow
Excited to release a huge evolution project on the works for many years: An experimental evolution project across climates to understand rapid adaptation Preprint: doi.org/10.1101/2025.0… All data available: grene-net.org/data #MOILAB @UCBerkeley @HHMINEWS 🧵👇
Rising Stars in Plant Sciences 2025 (RSPS2025) | Finalist Oral Presentation Contest | Jun 4 12:55-15:30pm & Jun 5 13:00-15:35pm (GMT/UTC) | FREE to Watch the Live Streaming via YouTube (youtube.com/@molecularplan…) or WeChat (MolPlant Channel)
Excited to share my new Tansley Review @NewPhyt on emerging single-cell and spatial omics technologies, many of which are just beginning to be applied in plant biology! Big opportunities lie ahead for the field. [1/n] nph.onlinelibrary.wiley.com/doi/10.1111/np…
Big leap forward in GRN inference by @manusaraswat10 & a great example of how deep learning models can reveal fabulous biology when developers really make an effort to design them and use them for discovery.
🧠 Excited to share my main PhD project! We mapped the regulatory rules governing Glioblastoma plasticity using single-cell multi-omics and deep learning. This work is part of a two-paper series with @bayraktar_lab , @OliverStegle and @MoritzMall groups. Preprint at end🧵👇
We're excited to share another work from our lab this week. We present ENTRAP-seq, the first massively parallel reporter assay to characterize protein-coding libraries in plants. Congrats to Simon, Lucas, and all other co-authors. biorxiv.org/content/10.110…
Glad to see this out! The ability to map protein footprints with high resolution has been missing in the field, but no longer. Finally some definitive answers on the regulatory potential of maize transposons. Fantastic work by @RNAnerd and co. nature.com/articles/s4147…

Happy to share our new publication out today in @NaturePlants where we quantitatively dissect synergy and antagonism that occurs during Nicotiana benthamiana transient agroinfiltration. Congrats to Simon, Matthew, Mitch and all other co-authors! nature.com/articles/s4147…
Happy to share a preprint describing my new method, Dam-IT (DamID-seq Incorporating Transcriptomics)! Dam-IT simultaneously captures TF-DNA binding, gene regulation, and chromatin accessibility from the same batch of plant cells. biorxiv.org/content/10.110…