Guignard Lab
@GuignardLab
Computer Science, Morphogenesis & Variability: From developing code to decoding development. @IBDMmarseille Open positions: http://www.guignardlab.com/jobs
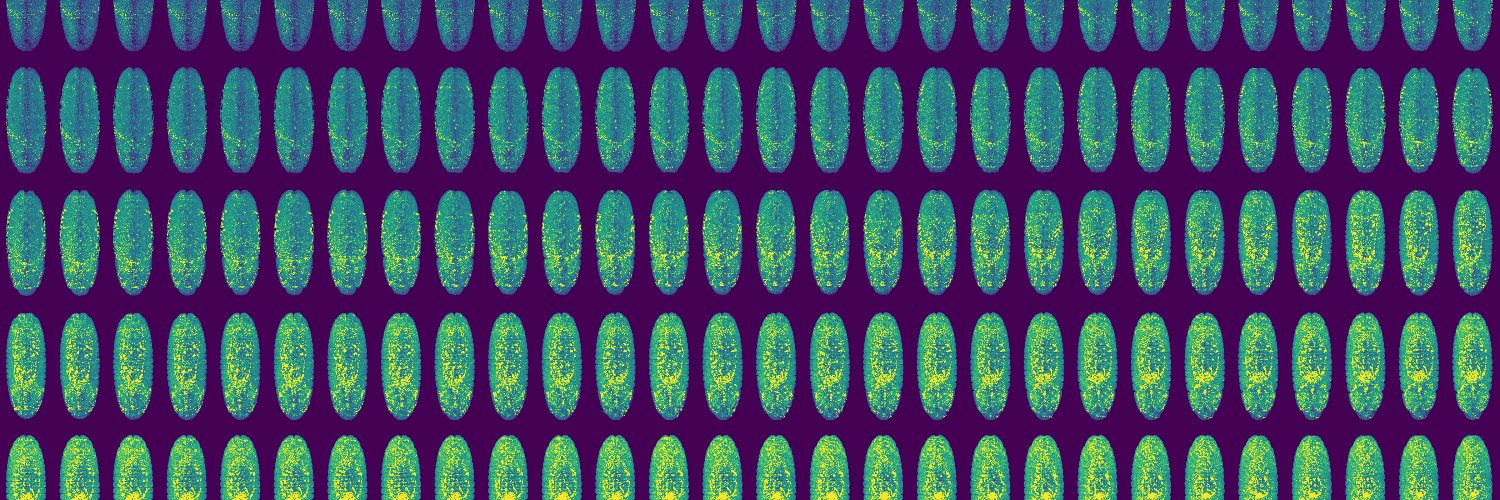
🚨📢Our paper on imaging, segmenting, visualising and measuring patterns in gastruloids across spatial scales w/ @napari_imaging is out‼️ An project led by @_AliceGros, @JVanaret, @DunsingValentin et al.🤓 A collab. between @Equipe_lenne, @PhilippeRoudot's and our lab. ⬇️Link⬇️
#Gastruloids have many uses as #models for: early mammalian development, #SelfOrganization, mapping signalling/gene expression, morphogenesis. Here @Equipe_lenne use’m to develop gral tools to image gene/protein expression at diff scales biorxiv.org/content/10.110… #InNumbersWeTrust
Delighted to share our latest work on #Ovulation, now published in @NatureCellBio! 🎉 Grateful to my teammates @Tabea_Marx and @sarahpenir in @SchuhLab at @mpi_nat for excellent teamwork. 🙌 Dive into the details here 👇 #Ovary #Egg #Reproduction #CellBiology #Fertility
Each month an #egg is ovulated from the #ovary, starting the journey of reproduction. But how does ovulation occur?🥚 Our latest work in @NatureCellBio led by @LastChrisThomas and @Tabea_Marx, describes the control of #ovulation using live imaging. bit.ly/3Ya8dEL (1/7)
Our paper ‘Whole-embryo Spatial Transcriptomics at Subcellular Resolution from Gastrulation to Organogenesis’ is now on biorxiv! We introduce weMERFISH, a platform for profiling gene expression in entire embryos at subcellular resolution. Here is🧵biorxiv.org/content/10.110… (1/12)
🚨Preprint alert🚨 Excited to share the #preprint version of our work @MundlosLab ! Curious about how a transposable element (TE) insertion can cause developmental phenotypes? 🧬🦠🐁 Check the 🧵 below and the full version on BiorXiv: biorxiv.org/content/10.110…
1/ I'd like to share a short thread about our project to build a nationwide, distributed core-facility for bioimage analysis. We discuss this in our recent preprint: arxiv.org/abs/2409.15009
🚀Applications are open for the 2025 edition of our EMBL Deep learning course for microscopy image analysis. This time, we will focus on more advanced methods so if so if you already know how to train models you will be in for a treat! Deadline Nov 25th
Are you ready to apply deep learning-based methods to your data and image analysis problems? If you've already trained a network in your own code, this is for you! #EMBLDeepLearning 🗓️ 17 – 21 February 2025 📍 EMBL Heidelberg 📩 Apply by 25 November ➡️ s.embl.org/mac25-01
📢Our preprint list is now up on FocalPlane. This list is focussed on new research in bioimage analysis. Drop us a message if there is a preprint that we've missed! focalplane.biologists.com/2024/09/06/mic…
🚨Job alert! @PoLDresden offers excellent postdoc fellowships. It includes up to a 4-year position with salary and research funds. The next deadline for application is 01.11. Don't miss this opportunity! 🤓 Please RT #postdocjobs physics-of-life.tu-dresden.de/career-educati…
New preprint: We have developed an integrated pipeline to i) image gastruloids in toto with dual-view 2-photon🔬, ii) quantitatify cell and tissue-scale patterns of gene expression and morphometric features, iii) interact with the data in @napari_imaging. biorxiv.org/content/10.110…
I am so excited to share this part of my PhD where we tackle the question: What impact do 3D contacts have on how enhancers regulate their target genes? biorxiv.org/content/10.110… 1/
🚀 What an incredible 2024 hackathon! 🌟. Congrats to all teams for their innovative minds and hard work. See you next year!💻✨ #CENTURIHACK2024
Congrats to our winning team on the Deep Force project ✨💻 @felix_rico @ismahene_mesbah #CENTURIHACK2024
From our speaker: “like my mom said, when things go wrong, you can always use a Fourier transform” #CENTURIHACK2024
Présentations are starting at #CENTURIHACK2024 ! Impressive results!



