
Gerstein Lab | Yale
@GersteinLab
Research in #Biomedical #DataScience & #Bioinformatics #CompBio. @MarkGerstein AT @YaleMBB @YaleMed
New study conducted by our lab in collaboration with #PsychENCODE has made significant discoveries linking genetic variants to genes and cell types in human brain. #psychencode24 For more details, refer to our original thread: x.com/MarkGerstein/s… news.yale.edu/2024/05/23/tra…
New paper on single-cell genomics & regulatory networks for 388 human brains just out in @ScienceMagazine. Neat stuff on single-cell QTLs, cell-to-cell communication, & DL models simulating drug effects (science.org/doi/10.1126/sc…) #PsychENCODE24
Excited to share our new paper in @CellCellPress on Digital Phenotyping from Wearable Biosensors using AI to characterize Psychiatric Disorders & identify Genetic Associations (led by @JasonJLiu & @beaborsari) doi.org/10.1016/j.cell…
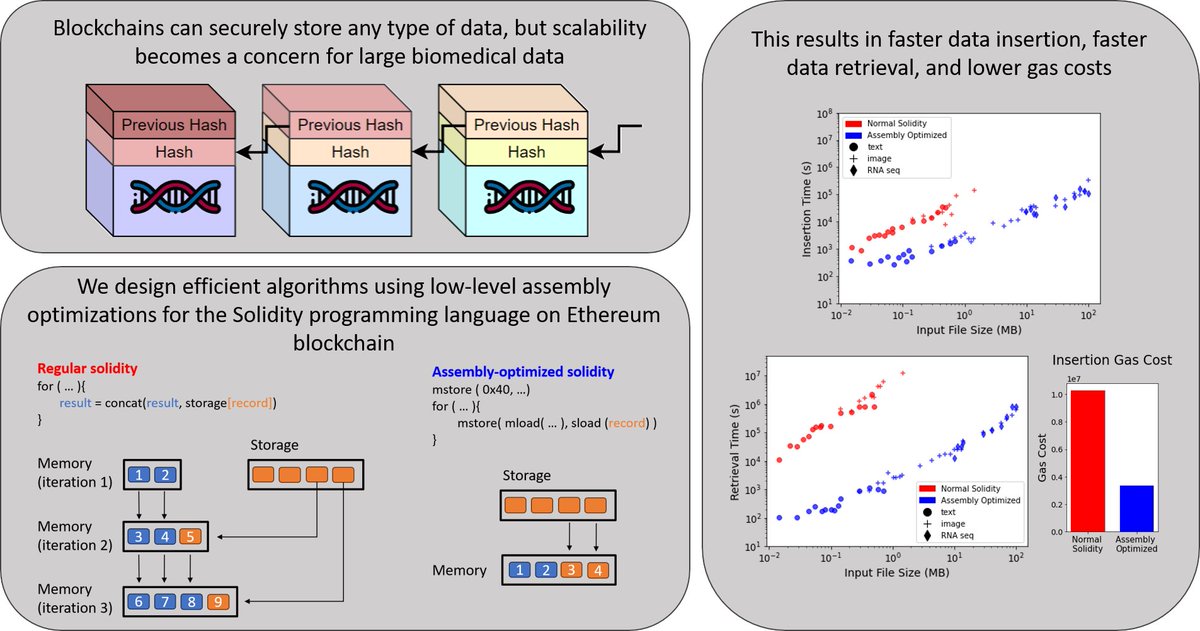
Our new paper in @jbi_journal describes the iDASH-winning method for efficient blockchain storage of biomedical data. We cut gas costs by 60% and sped up retrieval 500x with low-level Solidity optimization. By Eric Ni, Elizabeth Knight, @MarkGerstein doi.org/10.1016/j.jbi.…
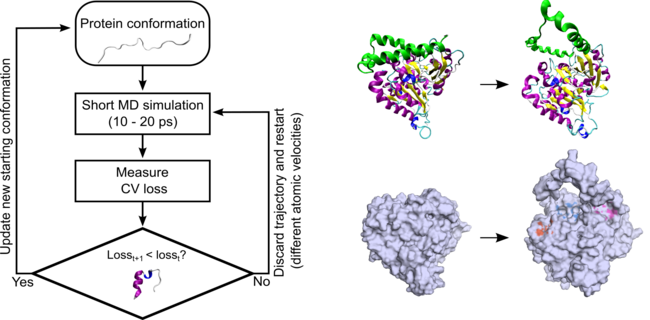
New @BiophysJ paper by Alan Ianeselli, Joe Howard and @MarkGerstein . A Molecular Dynamics algorithm to rapidly compute protein folding pathways and identify folding intermediates for targeted drug discovery! doi.org/10.1016/j.bpj.…
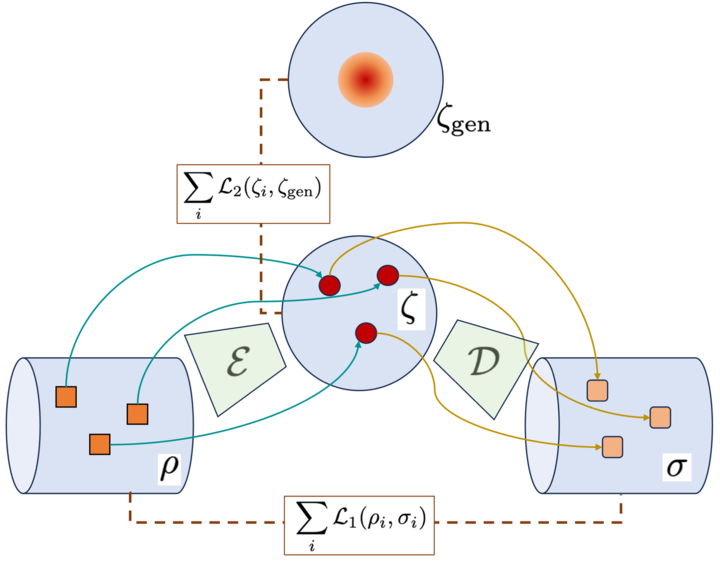
New @PhysRevA paper by Gaoyuan Wang, Jonathan Warrell, Prashant Emani and @MarkGerstein ! Check out our new model QVAE, a fully quantum variational autoencoder with latent regularization: journals.aps.org/pra/abstract/1…
I am very excited to share that our(@GersteinLab) paper was recently published in Cell! 🎉☺️ doi.org/10.1016/j.cell… had a lot of fun creating two versions of covert art for our paper: which one do you like more?
🚨Alert: Funded postdoctoral position available in our lab!
Just found out that we have an immediate postdoc opening for US nationals (citizens/green-card holders). Needs to be filled within 6 months. Lots of fun topics (e.g. biosensors, brain genomics, AI for bio, &c). If interested, see jobs.gersteinlab.org & contact me.
New @PLOSONE paper by Xiao Zhou, Sanchita Kedia, Ran Meng, and @MarkGerstein. Our deep learning framework analyzes fMRI scans for early Alzheimer's Disease detection, achieving 92.8% accuracy with a focus on model interpretability: doi.org/10.1371/journa…

New @CellRepPhysSci paper by Gaoyuan Wang, Jonathan Warrell, Suchen Zheng and @MarkGerstein! Check out our GP-GNN framework that jointly learns graph structure and node representation, with the two optimization procedures dynamically informing each other: cell.com/cell-reports-p…

Improved Prediction of Ligand–Protein Binding Affinities by Meta-modeling • This study introduces a meta-modeling framework integrating classical docking scores and sequence-based deep learning (DL) predictions to enhance ligand-protein binding affinity predictions. • The…
REliable PIcking by Consensus (REPIC) is now published in @CommsBio 🎉 REPIC harnesses multiple #cryoEM particle pickers to improve #particlepicking, achieving expert-level results with minimal manual intervention. Check it out! doi.org/10.1038/s42003… #research
Posting my keynote tomorrow at the @YaleMed Single-Cell Symposium, featuring lots of work by @YaleCBB students lectures.gersteinlab.org/summary/Large-… Large-scale analysis of Brain Single-Cell Data @YaleData