Pranam Chatterjee
@pranamanam
Designing biologics to program biology! 💻🧫 Assistant Professor at @Penn | Co-Founder @GametoGen and @UbiquiTxINC | Formerly @MIT '16 '18 '20 and @harvardmed
The next chapter of my lab's story begins today at @Penn as I join the faculty of @pennbioeng and @CIS_Penn! ❤️💙 My wonderful students and I will continue to push the frontiers of generative algorithm theory to design and translate next-gen biologics for unmet/rare indications…


Toxic heavy metals contaminate ecosystems and harm vulnerable communities. 🌎⚠️ Here, we present Metalorian, a conditional diffusion model that generates heavy metal-binding peptides, validated by MD and experiments! 💻🧪 📜: biorxiv.org/content/10.110… 🤗: huggingface.co/ChatterjeeLab/……

Most biological processes, like stem cell differentiation, branch into multiple fates, but current trajectory inference methods only predict single paths.🌳To fix this, we introduce BranchSBM 🌿, from our unstoppable @_sophia_tang_ ! 🌟 📜: arxiv.org/abs/2506.09007 💻:…
Easily one of the coolest advances of the year! 🔥 A one-step normalizing flow training method with no expensive Jacobian computations? Imagine how efficient sampling of complex bio distributions will be now! 💨 Congrats @AlexanderTong7 @bose_joey @mmbronstein + team! 🙌
Excited to release FORT, a new regression-based approach for training normalizing flows 🔥! 🔗 Paper available here: arxiv.org/abs/2506.01158 New paper w/ @osclsd @jiarlu @tangjianpku @mmbronstein @Yoshua_Bengio @AlexanderTong7 @bose_joey 🧵1/6
I absolutely love this paper!! 🌟 Beautiful work by @jsunn_y @yisongyue and team to show that discrete diffusion models can be effectively guided by fitness data from low-throughput wet-lab assays! 🧪 I'm telling you guys: guided sequence generation is where it's at! 🤙
I'm excited to share our new preprint "Steering Generative Models with Experimental Data for Protein Fitness Optimization" (🧵1/5)! Paper: arxiv.org/abs/2505.15093 Code: github.com/jsunn-y/SGPO
As you know, we’ve been deep in the discrete diffusion trenches for sequence design for quite some time now (both theoretically and for biology!)—PepTune for multi-objective discrete diffusion for therapeutic peptide design from @_sophia_tang_, P2 for path planning and improved…
We’ve developed Gemini Diffusion: our state-of-the-art text diffusion model. Instead of predicting text directly, it learns to generate outputs by refining noise, step-by-step. This helps it excel at coding and math, where it can iterate over solutions quickly. #GoogleIO
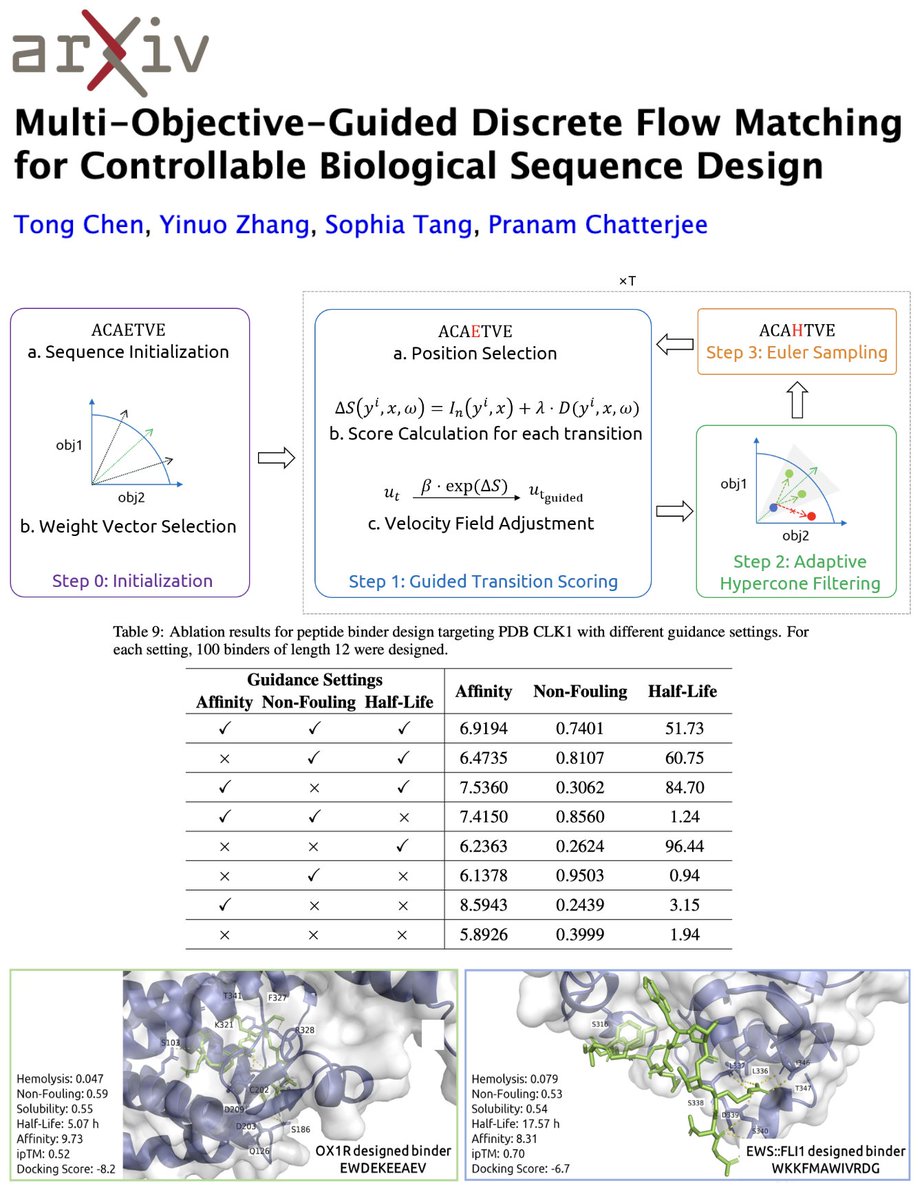
Biologics need optimization across multiple properties—affinity, half-life, non-fouling, etc. 💊 PepTune did this with discrete diffusion + MCTS 🌳, but flow matching may give us smoother, more structured control. 🌊 Meet MOG-DFM! 🌟 📜: arxiv.org/abs/2505.07086 💻:…
This was one of my favorite experimental papers from our @gembioworkshop at ICLR!! 🇸🇬 A super crafty way of generating longer DNA libraries via Golden Gate -- we'll definitely be using it for our proteome editing screens!! 🧬 Really great work @ShaozhongZou!! 👏
🚀 I’m delighted to share the first public release of our work on OpenReview! Starting from @TwistBioscience 300 bp oligo-pools, we assembled longer DNA libraries (~600 bp), containing up to 96 gene-sized sequences, each constructed from 4 oligos. 🧬
What a way to end #ICLR2025 in Singapore 🇸🇬 with an acceptance for PepTune in #ICML2025 in Vancouver 🇨🇦! 🥳 So many congratulations to my brilliant students @_sophia_tang_ and @yinuo_z98! 🫶 A huge moment for our lab! 🙌 📜: arxiv.org/abs/2412.17780 A quick TLDR for PepTune, if…
So I know this is Christmas Eve🎄, but I am so excited to share this INCREDIBLE breakthrough algorithm from my lab! I promise I'm not exaggerating: let me explain why! 😉 I'm sure you guys all know about Ozempic and Wegovy by now? 😅 GLP-1R agonist peptides, which when…
Super excited to be at #ICLR2025 in Singapore! 🇸🇬 My students and I are presenting 20+ accepted works across the workshops/main, including at @ai4na_workshop @lmrl_bio #DeLTa #FPI! We're especially excited to co-host the @gembioworkshop tomorrow!! 💻🧬🧫 Come say hi!! 👋

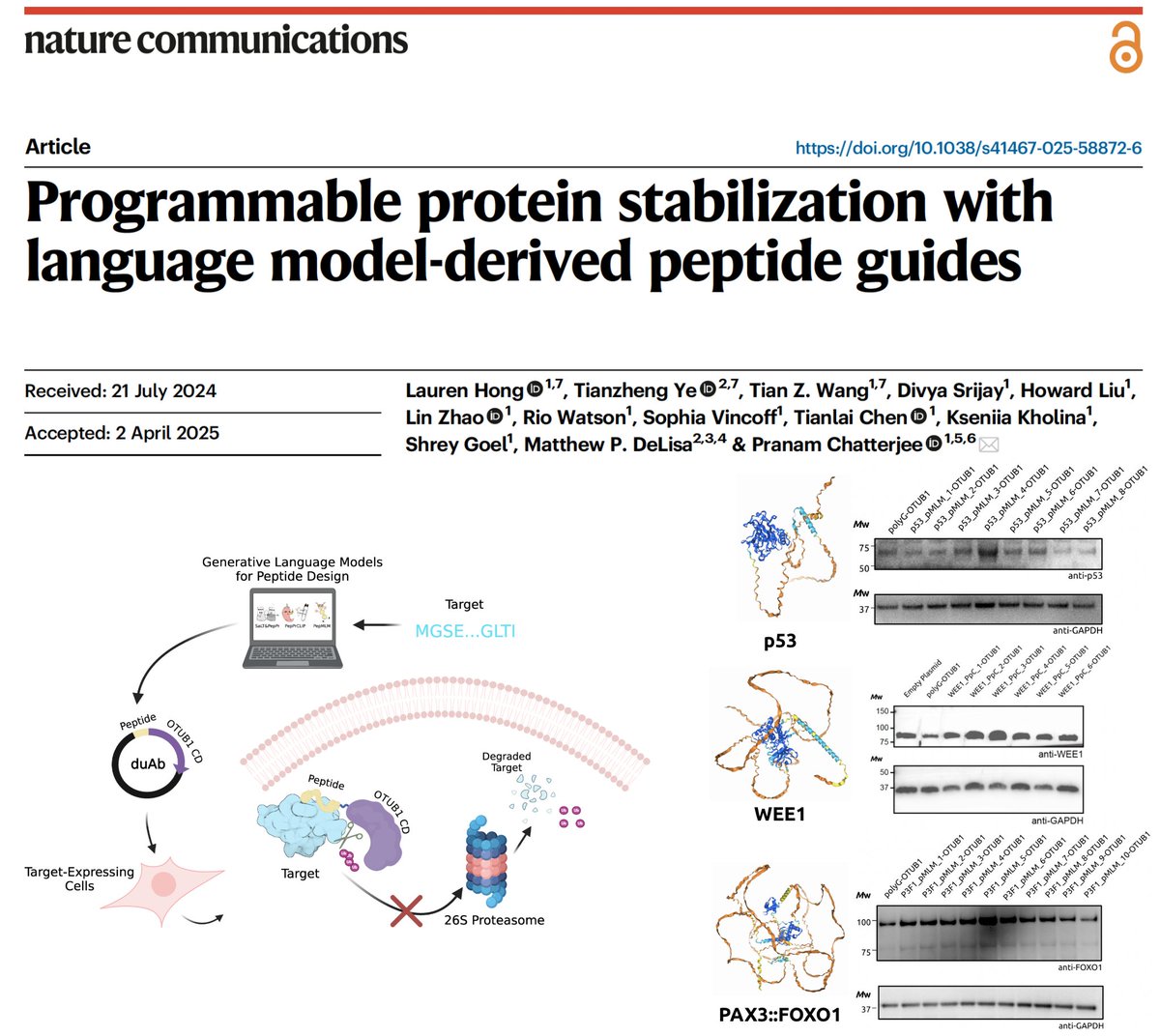
Our pLM-designed peptides, when attached to E3 ligases (aka uAbs), enable us to programmably degrade disease targets (think CRISPRi, but for proteins). But what if we want to STABILIZE useful proteins? In @NatureComms, we introduce duAbs! 🌟 📜: nature.com/articles/s4146… 🧬:…
PTM-Mamba is out in @naturemethods!! 🥳 PTM-Mamba is the first/only pLM to encode both WT and post-translationally modified residues as tokens! And we do this without losing any base protein information from ESM-2! 🤝 📜: nature.com/articles/s4159… 💻: github.com/programmablebi… We…