OpenProtein.AI
@openprotein
Boltz-1 & Boltz-2 now live via GUI & APIs! Predict protein, protein–RNA/DNA/ligand structures with confidence scores & binding affinity metrics for virtual screening. Compare finetuned models in the new overview page to find your best performer fast. openprotein.ai/early-access-f…
Product update: Indel Analysis lets you score insertions/deletions across your sequence using PoET-2. You can now also compare multiple 3D structures in Mol* to evaluate design alternatives. Sign up now: openprotein.ai/early-access-f…
Why does no one in AI protein engineering work on indels? We’re solving this at OpenProtein.AI. Check out our upcoming indel design tooI! 🤩 1/4
Product update: PoET-2 now supports structure inputs for enhanced prediction and design via Python APIs. Check out our new inverse folding tutorial to see it in action. 🔗 docs.openprotein.ai/walkthroughs/P… Sign up for OpenProtein.AI: openprotein.ai/early-access-f…

🧬 Protein Revolution: The Tiny Model Making a Massive Impact! PoET-2 is changing the game in computational protein design, slashing experimental data needs by 30x! 🚀 learn more: synbiobeta.com/read/protein-r… #ProteinDesign #BiotechInnovation #AIRevolution
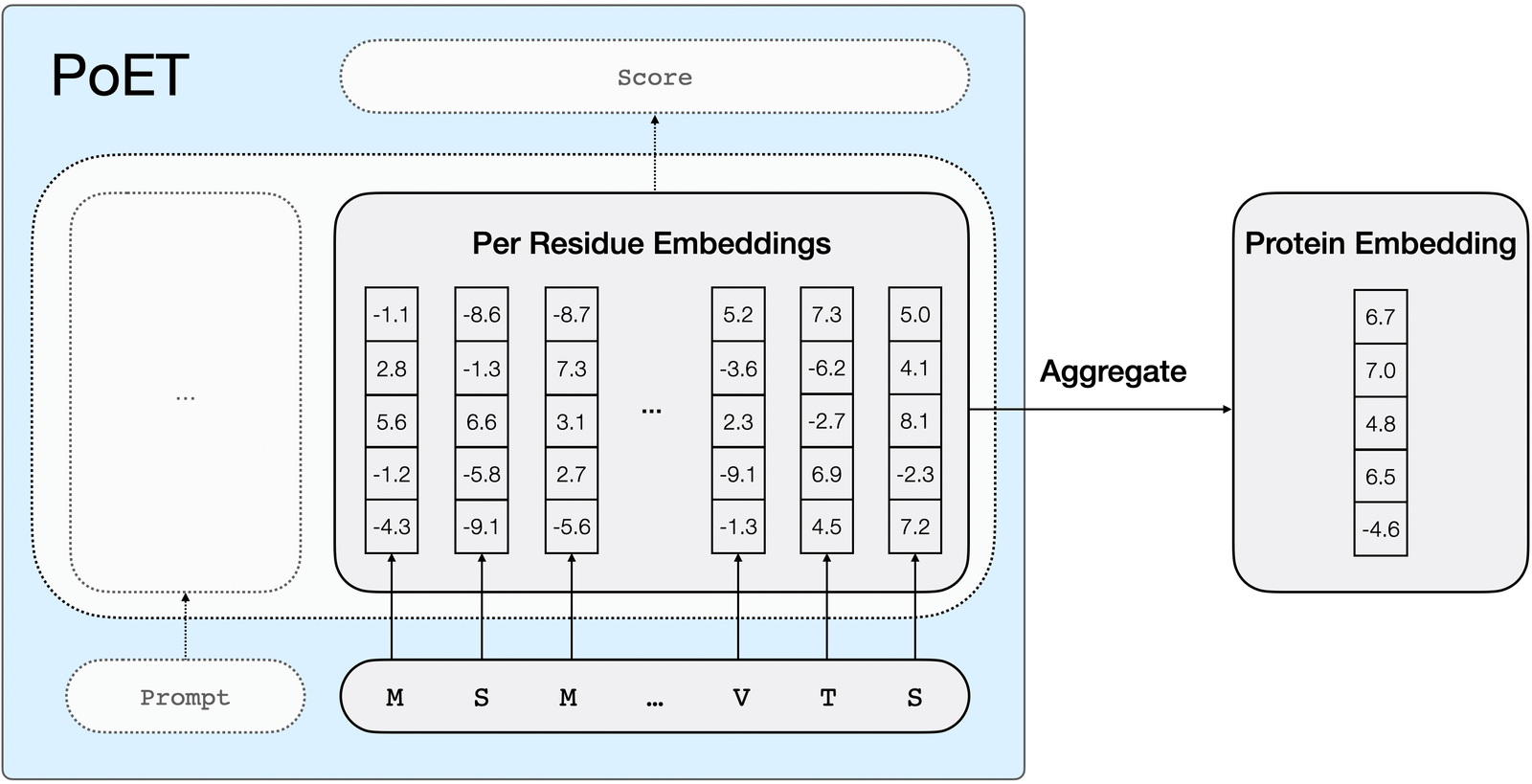
New blog post! PoET, our generative protein language model, is also super powerful for fine-tuning. PoET outperforms previous methods for supervised variant effect prediction even with 15x less data. openprotein.ai/poet-foundatio…
Engineering protein complexes or multimeric proteins is challenging! Our platform lets you model critical interactions between subunits easily. See our latest walkthrough for an example of how to design optimized variants with cross-chain mutations. docs.openprotein.ai/walkthroughs/m…