Open-ST
@open_sts
Open-Source Spatial Transcriptomics developed in the @N_Rajewsky lab, @MDC_Berlin
Terrific work, huge congrats Marie, Daniel, Elena, Pino, Nikos, Nikolaus and everyone involved!! Likely a game changer for many labs doing spatial omics 🚀🚀🚀
Our #OpenST paper is now online @CellCellPress! authors.elsevier.com/c/1jJckL7PXqR3U Very cool updates ahead🧵⬇️ @L_Marie_Schott @_Elena_93 #GiuseppeMacino @nu_karaiskos @N_Rajewsky
In the latest issue! Open-ST: High-resolution spatial transcriptomics in 3D dlvr.it/TBB6Kl
The @N_Rajewsky lab at the #mdcBerlin has developed a spatial transcriptomics platform to reconstruct tissues in #3D with subcellular resolution. The platform will enable a deeper understanding of basic biology and of how diseases develop: mdc-berlin.de/news/press/3d-… @CellCellPress
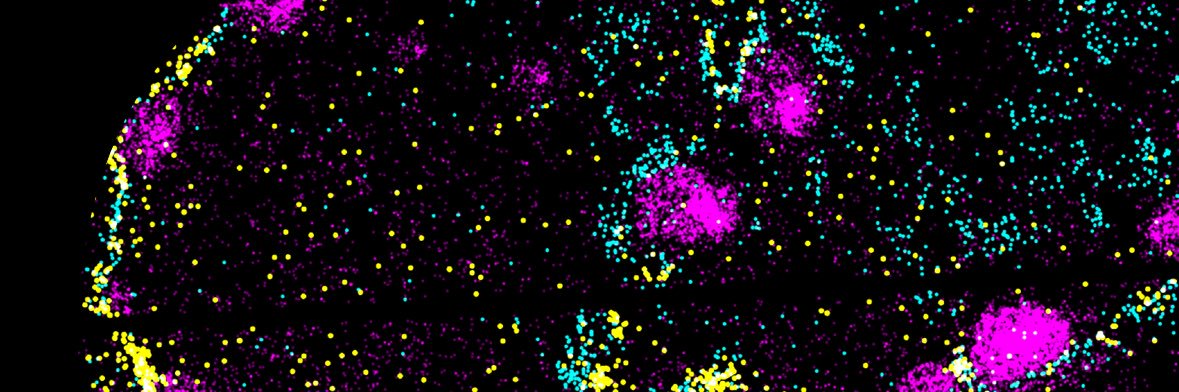
i am so excited about our open source inexpensive, sub-cellular capture, do-it-yourself Open-ST that transforms tissues into virtual tissue blocks in 3D - IDing pathways & gene programs driving phenotypes. a huge congratz to the team! cell.com/cell/abstract/…
And it's out 🎉 Check out our updated paper & tweetorial on Open-ST, our DIY spatial transcriptomics method. Many, many thanks to everyone involved in this huge & exciting project! #OpenST #SpatialTranscriptomics
Our #OpenST paper is now online @CellCellPress! authors.elsevier.com/c/1jJckL7PXqR3U Very cool updates ahead🧵⬇️ @L_Marie_Schott @_Elena_93 #GiuseppeMacino @nu_karaiskos @N_Rajewsky
Our #OpenST paper is now online @CellCellPress! authors.elsevier.com/c/1jJckL7PXqR3U Very cool updates ahead🧵⬇️ @L_Marie_Schott @_Elena_93 #GiuseppeMacino @nu_karaiskos @N_Rajewsky
A spatial transcriptomics method that… 🧪is easy to use 🔬has high resolution 🪙is cost-efficient 🌍scales to 3D Just in time for Xmas: Open-ST, a end-to-end, open-source method that checks all boxes, from the @N_Rajewsky lab 📄Preprint: tiny.cc/openst 🧵⬇️