
luciedequiedt.bsky.social
@luciedqt
PhD student @JohnsHopkins. @ULiegeFSA Biomedical Engineering alumn. 🇧🇪 in the 🇺🇸 and waffle connoisseur.
Our kidney work being finally out, that means I get to share some of my favorite images 🤩 Starting off with the results of our deep learning segmentation. We can label 17 micro anatomical components of the fetal kidney on H&E images.

It’s #WorldKidneyDay! A day to bring attention to this incredibly important organ and #KidneyHealthForAll. Sharing this image of a kidney showing developing nephrons in magenta and the collecting duct system and part of the nephron in yellow. 🫘
📢 Excited to announce our latest pre-print: “Modeling dynamic social vision highlights gaps between deep learning and humans”! 🌟 w/ @emaliemcmahon, Colin Conwell, @michaelfbonner, and @leylaisi🧵 #NeuroAI #CognitiveScience [1/7]
New paper where we integrate 3D histology (CODA) with IHC and imaging mass cytometry (IMC) to produce 3D single-cell immune maps in the vicinity of pancreatic precursor lesions (PanINs). Read about CODA+IHC/IMC = 3D immune maps here: biorxiv.org/cgi/content/sh…
New Preprint from the lab: Single-cell morphology encodes functional subtypes of senescence in aging human dermal fibroblasts. We are super excited to share this one! biorxiv.org/content/10.110…
So excited I got to give my first talk yesterday, and I am so happy it was at #DDW2024! 🤩
Great talk today by @luciedqt at #DDW2024 on her work in early detection for individuals at high risk of development of pancreatic cancer!
Another paper out today in the American Journal of Surgical Pathology! Recently, we’ve shown that PanIN are more abundant and genetically diverse than previously thought. Here, we expand on this to show that 3D imaging challenges the 2D classification criteria of PanINs.
Sneak peek of some exciting work coming soon from the @JudeM_Phillip lab led by Pratik biorxiv.org/content/10.110…. Here is some drug clearance of senescent cells. We can segment, track, and analyze single cells through treatment, and gauge effectiveness through morphological change.
For the next 3 days, we are hosting the CCBIR/TEC conference at Hopkins. These two center networks are funded by the US National Cancer Institute. Warm welcome to all participants. CCBIR = Cancer Cell Biology Imaging Research TEC = Cancer Tissue Engineering Collaborative
I’m beyond excited to share work from my PhD, co-led with Alicia Braxton, out today in Nature! We used CODA to map the anatomic and genomic properties of pancreatic cancer precursors. Led by the amazing @lauradelongwood and @deniswirtz ! nature.com/articles/s4158…
CODA day! The CODA team at @JohnsHopkins maps pancreatic precursor lesions in 3D and at single-cell level. First 3D spatial genomic maps of large tissue samples. A human pancreas contains a staggering ~1000 lesions, some showing multifocality. Here: nature.com/articles/s4158…
Had a great time presenting our preliminary work on PanIN burden in high risk individuals yesterday at #USCAP2024 in Baltimore. Thanks to everyone who stopped by for the great discussions and to my mentor @AshleyKiemen for the support! 🤩
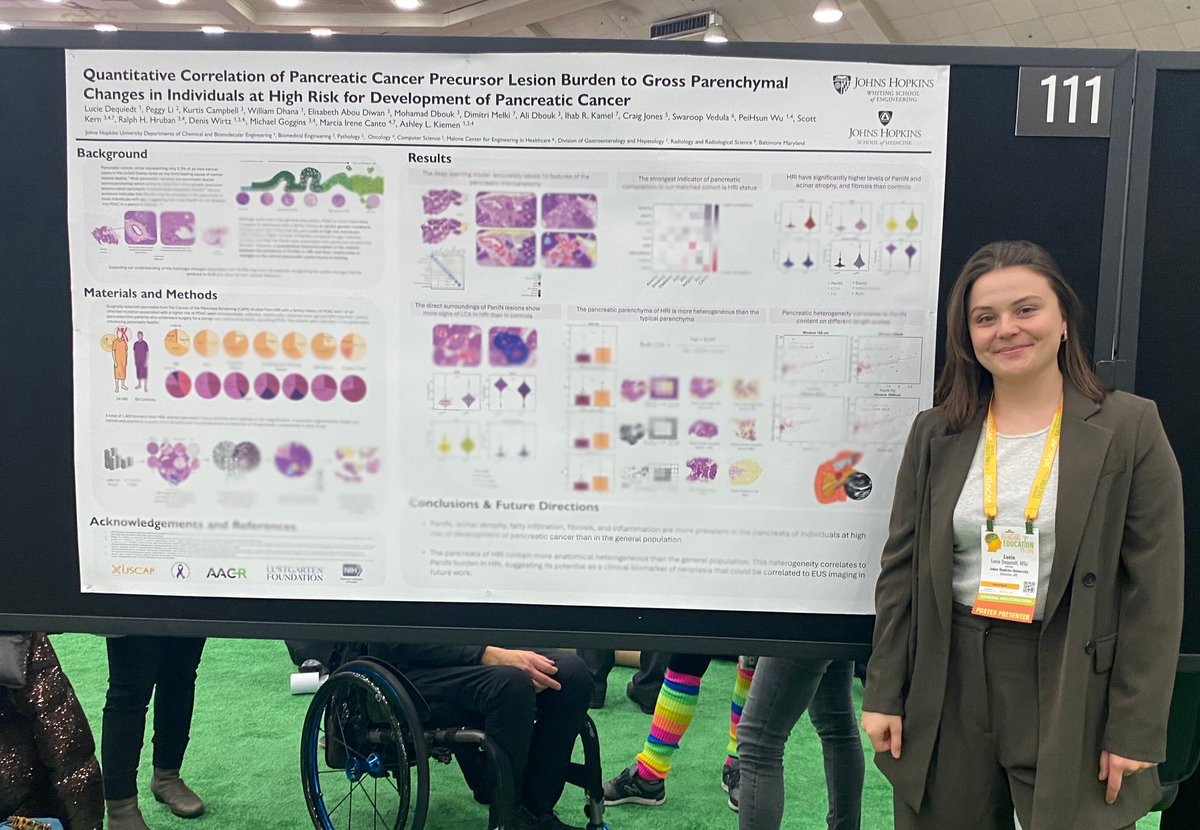
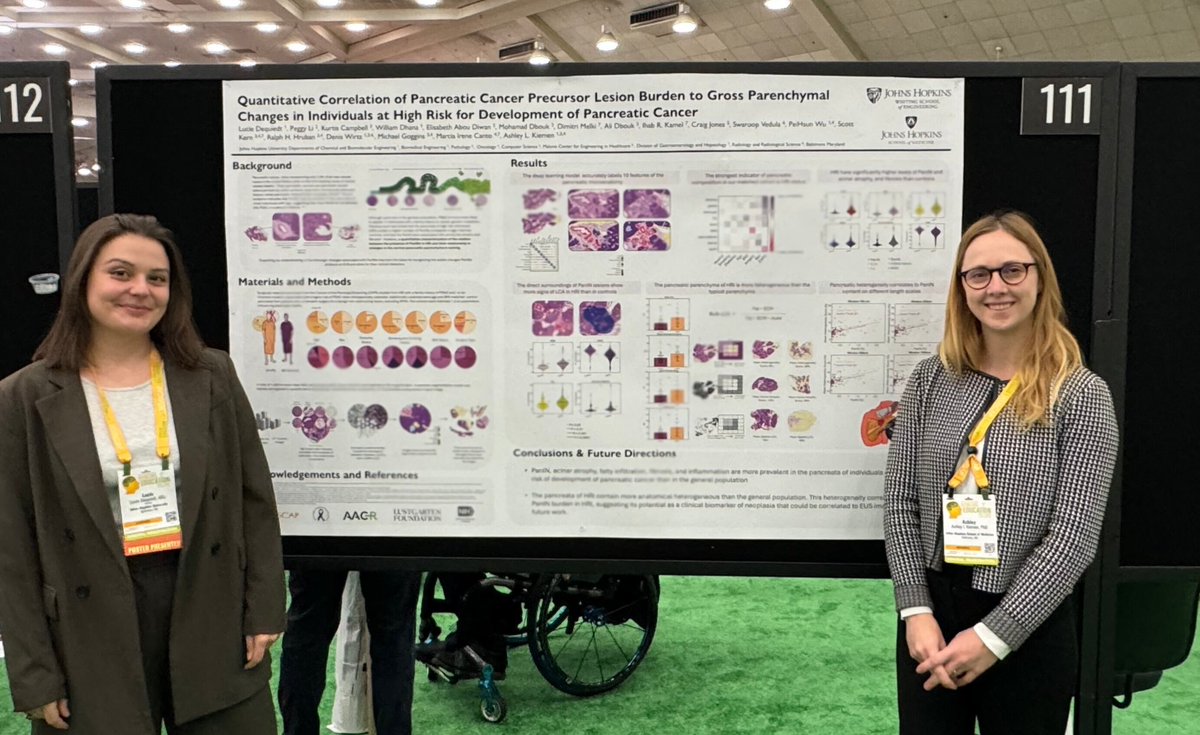
New paper out today! MRI-based measure of pancreatic fat correlates to histology-based measures of pancreatic fat, stroma, acinus, and PanIN content, especially in individuals at high-risk for development of pancreatic cancer. Read more here: journals.lww.com/pancreasjourna…
We wanted to start with a bang for our first paper in cell therapy and addressed the central problem with current CAR T cells: their poor infiltration in solid tumors. We created self-propelled CAR T cells...et voilà! Read here: biorxiv.org/content/10.110…
Celebrating the 1 year anniversary of the Kiemen lab tonight! It's been an incredible adventure and I'm excited for the many great things to come!! 🥰🥳