Kihara Laboratory
@kiharalab
Bioinformatics, protein modeling, cryoEM, drug screening, function prediction. Daisuke Kihara, professor of Biol/CS, Purdue U. YouTube: http://alturl.com/gxvah
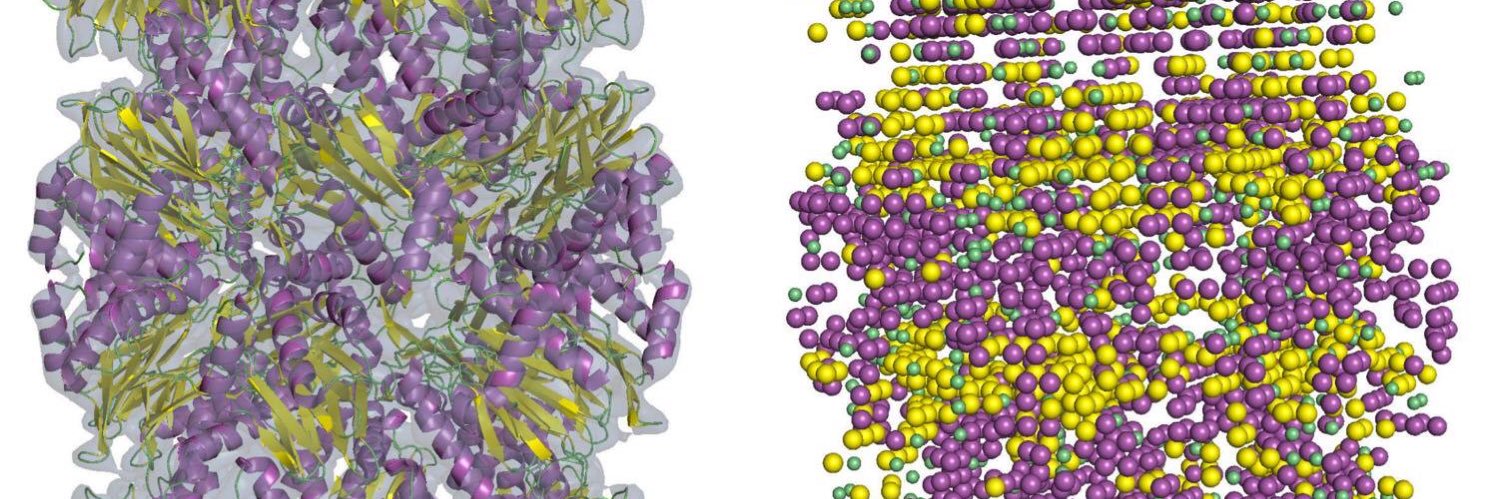
New paper accepted: "Structure Modeling Protocols for Protein Multimer and RNA in CASP16 with Enhanced MSAs, Model Ranking, and Deep Learning" Yuki Kagaya et al., Proteins. This reports our strategies in CASP. The initial submission available at ggnpreprints.authorea.com/doi/full/10.22…
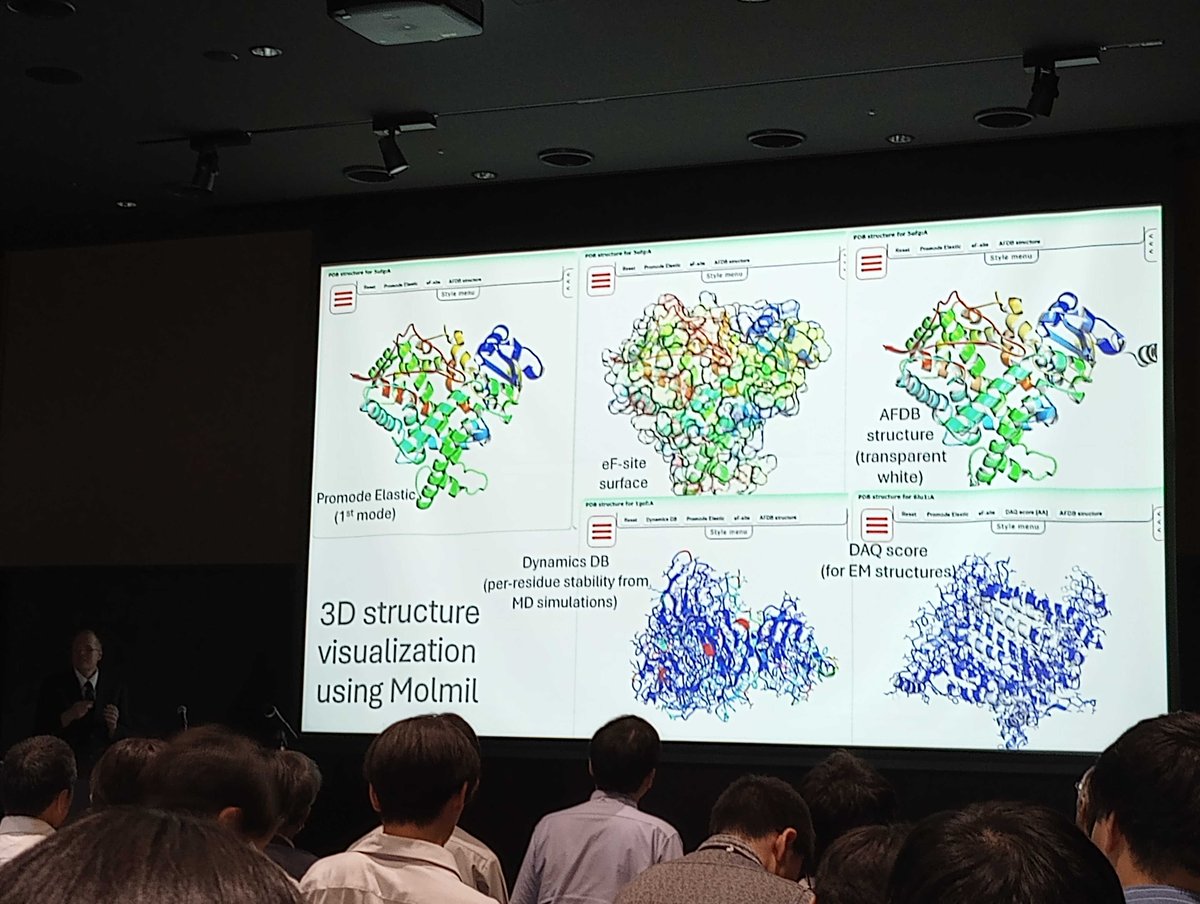
At ACA2025 in Chicago, I presented about our DAQ cryo-EM model quality score. アメリカ結晶学会で、電顕からの構造モデルのエラー評価スコア、DAQについて発表しました。ぜひ使ってみてください。 Available at em.kiharalab.org/algorithm/daqs… Quick YouTube tutorial: youtube.com/watch?v=YRAT98…
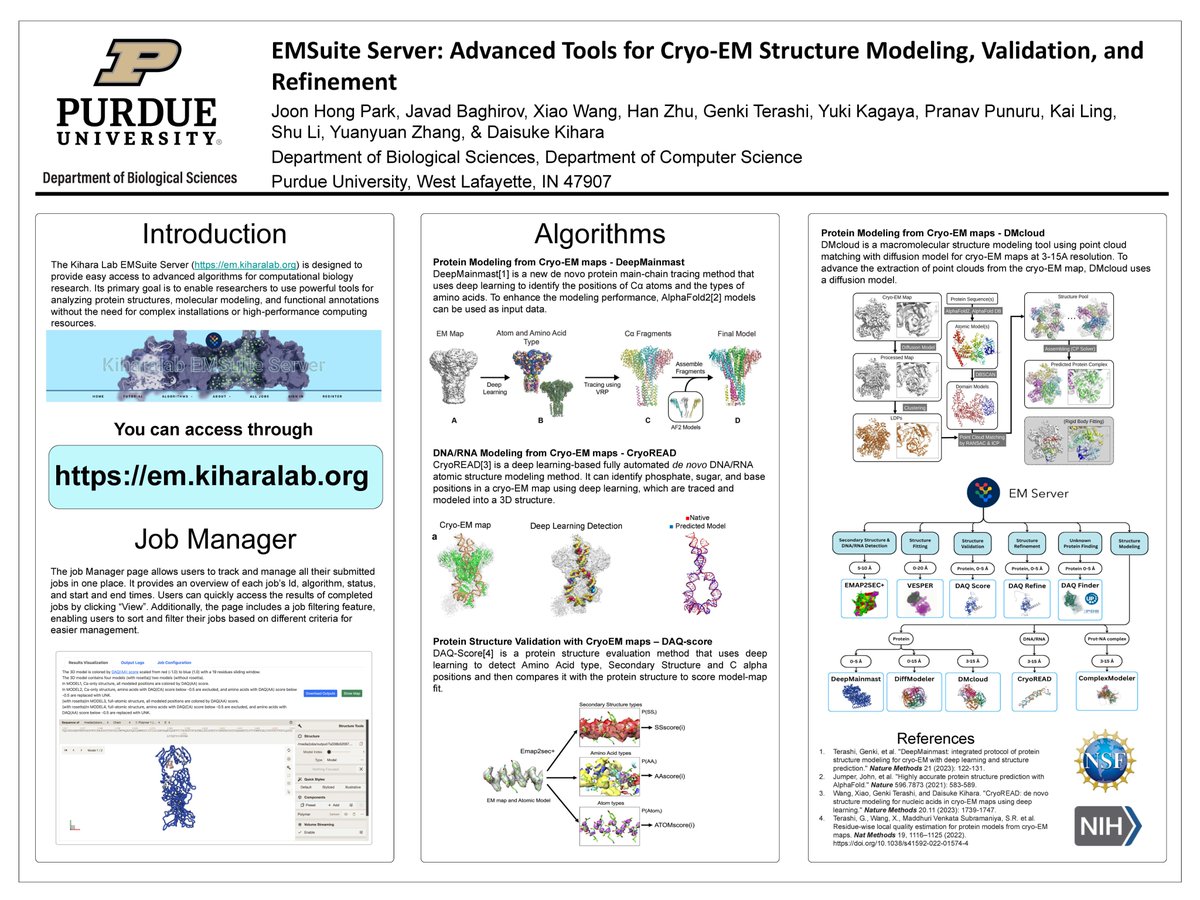
Joon Hong Park from @PurdueCS at American Crystallographic Association (ACA) 2025 meeting. presenting his poster about our cryo-EM structure modeling server. Visit the server for easy and accurate modeling at em.kiharalab.org
Dr. Eman Alnabati who did her PhD in our lab at @PurdueCS is featured by the Saudi Press Agency in a short video. She is now a postdoc at MIT. Great work, Eman!
#فيديو_واس | إيمان النباتي، باحثة ما بعد الدكتوراه في MIT ضمن زمالة “ابن خلدون”، تُطَوِّع الذكاء الاصطناعي لخدمة الرعاية الصحية، حيث تُركّز أبحاثها على تطوير تقنيات موثوقة تُسهم في تحسين جودة الخدمات الطبية. #واس_علمي
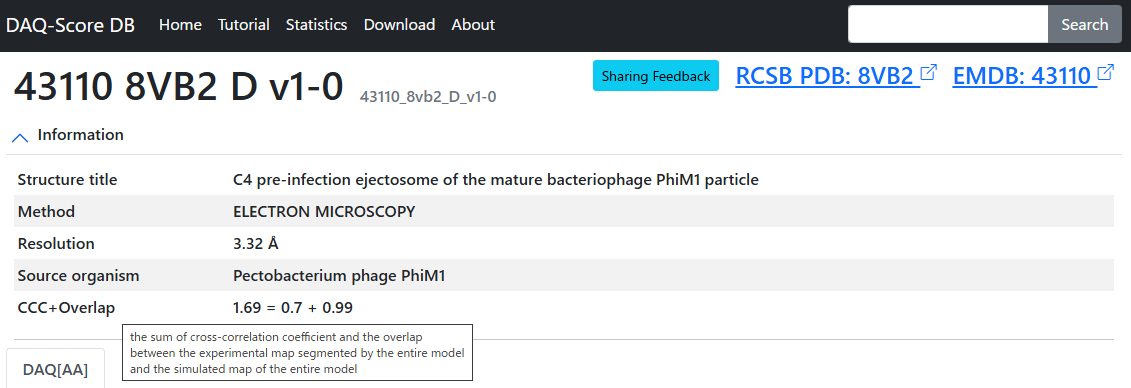
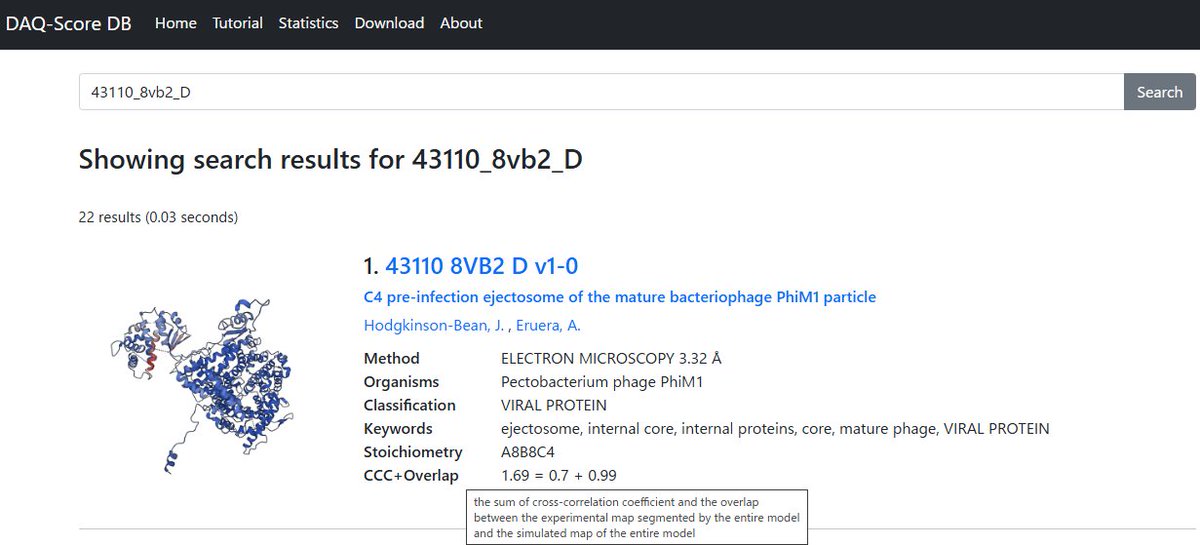
Happy July the 4th! DAQ-Score DB, cryo-EM model quality assessment DB is updated in the meantime - now for 246,454 PDB chains from 16,519 EM maps. Now Cross Corr + overlap value is provided as a str fitting metric. daqdb.kiharalab.org
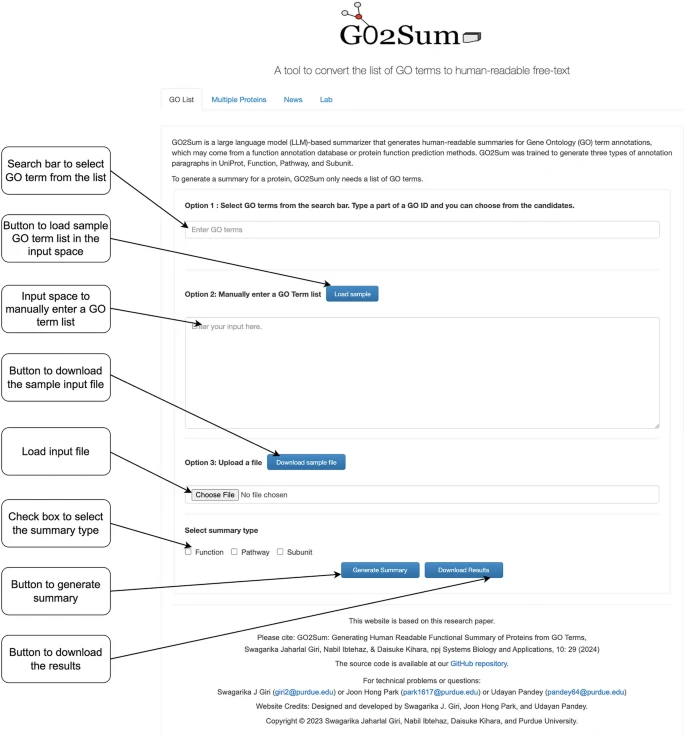
New book chapter online: Translating a GO Term List to Human Readable Function Description Using GO2Sum by @GiriSwagarika Udayan Pandey, @joonhongpark111 & @d_kihara Methods in Mol. Biol. The server is available kiharalab.org/go2sum_server/ link.springer.com/protocol/10.10…
Congratulations Emilia!!
Happy to share that I have successfully defended my MS thesis at Purdue University! “Assessing the Accuracy of Protein Function Prediction using Machine Learning” Grateful to Dr. Kihara and everyone in the lab @kiharalab
Happy to share that I have successfully defended my MS thesis at Purdue University! “Assessing the Accuracy of Protein Function Prediction using Machine Learning” Grateful to Dr. Kihara and everyone in the lab @kiharalab
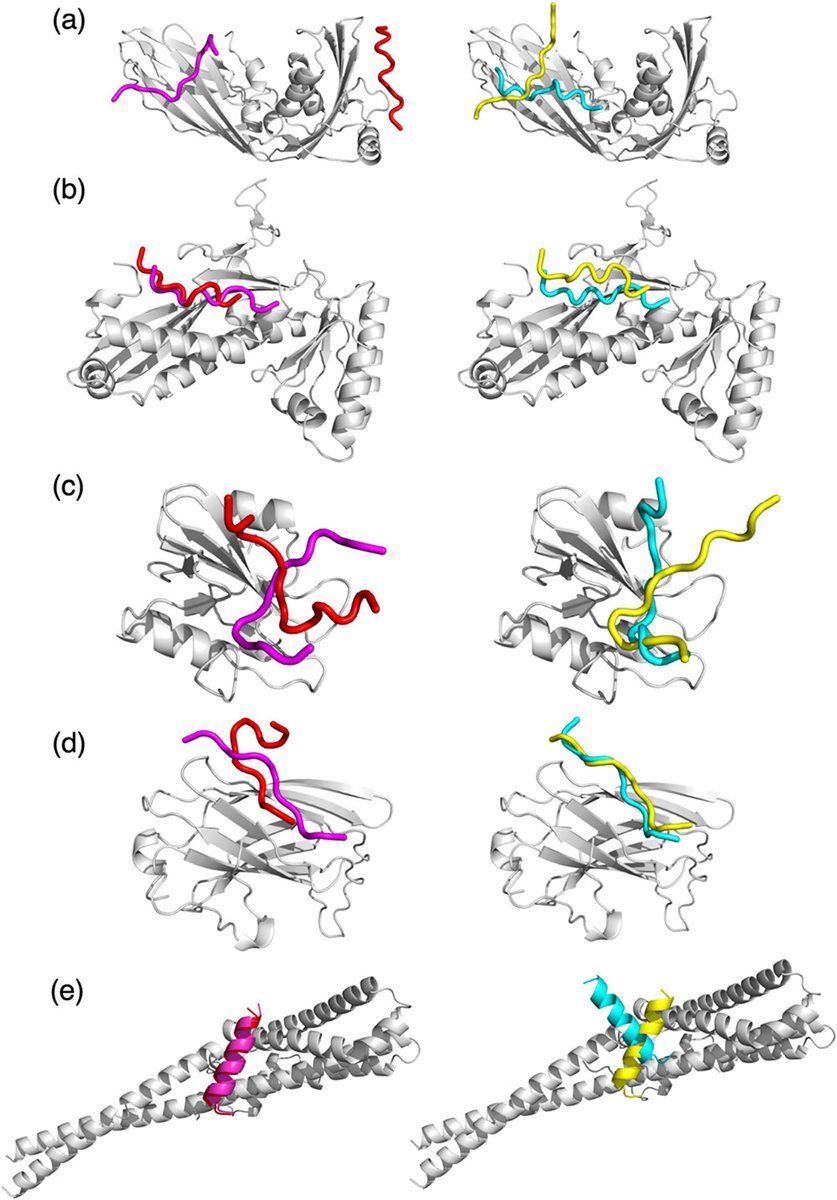
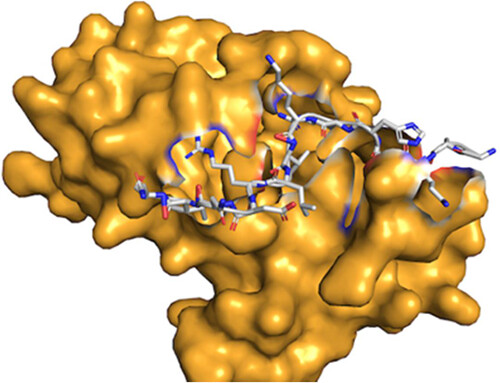
Distpepfold, our new peptide docking method is available as a source code and Google Colab notebook! Paper: pubs.acs.org/doi/10.1021/ac… Github: github.com/kiharalab/Dist… Google Colab: colab.research.google.com/drive/1Q1ecUy2…
Han Zhu from @purduecs presenting a collaboration project with KEK, Japan at the Annual Meeting of the Protein Science Society of Japan. "Automated Mask Creation and Class Selection in Single Particle Analysis".

DAQ cryo-EM structure validation score mentioned in the PDBj seminar at the Annual Mtg of Protein Science Society of Japan. You can select protein models from cryo-EM with sufficient DAQ in PDBj pdbj.org Quick intro of DAQ: youtube.com/watch?v=YRAT98…

🌀 Tertiary structure: 215 diverse 3D structures from recent PDB entries. NuFold leads monomers (0.393 TM-score), AlphaFold3 dominates complexes (0.381 TM-score). Non-Watson-Crick interactions remain a major challenge for all methods 6/9
Real-time structure search and structure classification for AlphaFold protein models nature.com/articles/s4200…
Domain-PFP allows protein function prediction using function-aware domain embedding representations nature.com/articles/s4200…
Researchers in Purdue’s College of Science have created a program named NuFold, which will help progress RNA research by over a decade. purdueexponent.org/campus/general…
New paper online : Learning with Privileged Knowledge Distillation for Improved Peptide–Protein Docking, ACS Omega. The method, Distpepfold, uses student-teacher model to improve peptide docking over AF2. pubs.acs.org/doi/10.1021/ac…

A recent paper on the RNA benchmark dataset, RNAGym, revealed that our NuFold outperforms other methods in predicting RNA 3D structures! Our NuFold is publicly available on our GitHub for everyone! NuFold paper: nature.com/articles/s4146… Code: github.com/kiharalab/NuFo…
Join our webinar on Cryo-EM modeling tools at SBgrid tomorrow (6/10), 12pm EST! "DiffModeler & CryoREAD: Macromolecular and Nucleic Acid Structure Modeling for Cryo-EM Maps", by @gterashi sbgrid.org/webinars/