
Brian Searle
@briansearle
Career Scientist in Quantitative Health Sciences at @MayoClinic and @proteome_sw into immuno-oncology, proteomics, mass spectrometry & bioinformatics (he/him)
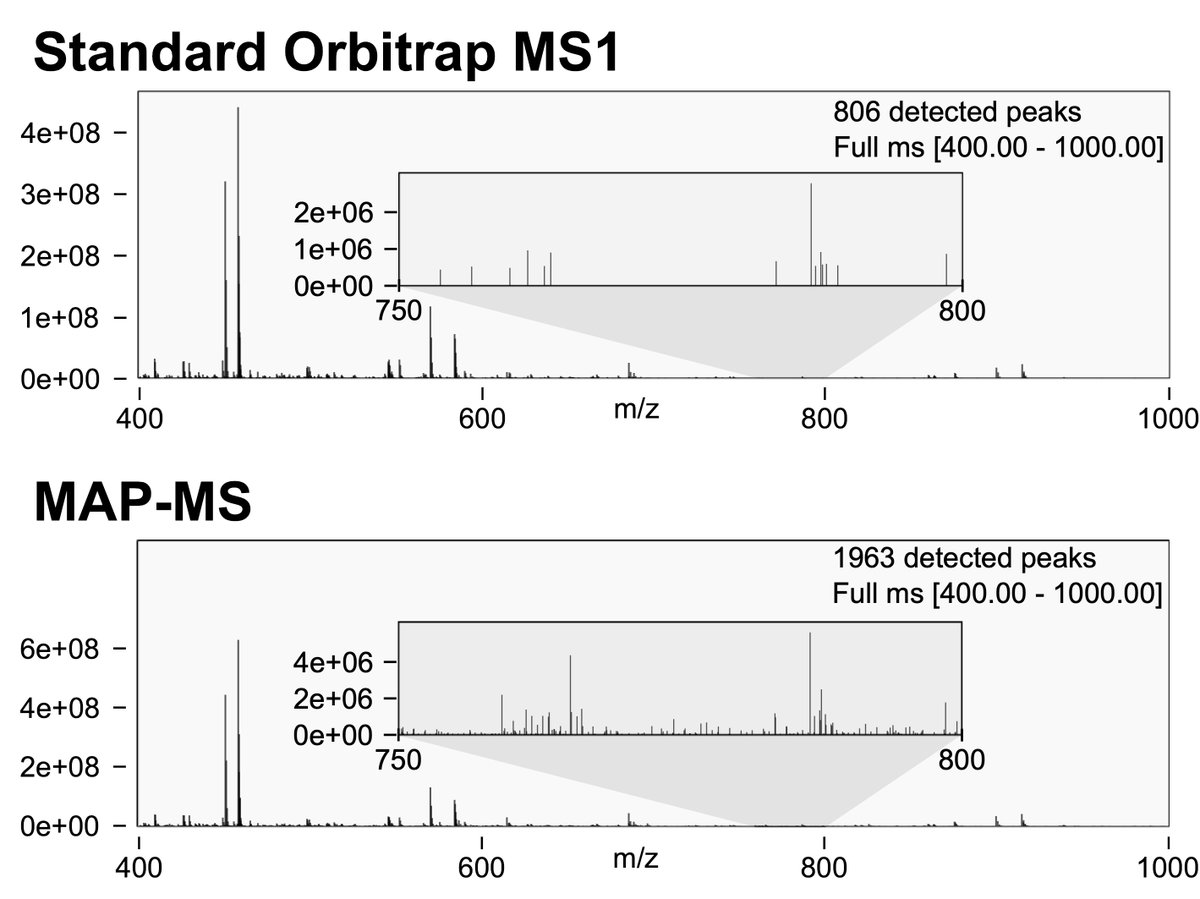
People have asked about how MAP-MS works so here's a tweetorial on @teerap16's new preprint: biorxiv.org/content/10.110…. MAP-MS takes advantage of parallelization in a trapping MS, collecting Orbitrap MS1s with 2x the dynamic range by differentially amplifying parts of the ion beam.
New research in @NatureComms on rapid assay development for low input targeted #proteomics using a versatile linear ion trap. This method promises to accelerate #immuno_oncology research with #targeted_proteomics using minimal sample input. @ArianaShannon @briansearle @Zihai…
So proud of @V_Akshintala well deserved! We are going to do great things for the diagnosis and management of #pancreatitis @briansearle .. be careful of the triple effect 😎
Congratulations to The NPF 2024 Pancreatitis Research Grant Awardee, Venkata Akshintala, MD!👏🏼 This technique demonstrates a huge advancement in the treatment and care of pancreatitis. @apapancreas @chirhoclin @HopkinsMedicine
@briansearle w/ @OSUWexMed presenting his work on urine proteomics as identifiers of chronic #pancreatitis biomarkers. His work, in collaboration w/ @MaisamHaijaMD and the PCC won the Best Abstract in #Pediatric #Pancreas #Research at #APA2024! @CincyKidsGastro
A wonderful lecture by @briansearle on #proteomic #applications in #childhood #pancreatitis, next phase #collaboration with @SanthiVegeMD will be great. #TeamworkMakesTheDreamWork @TMFoundationHQ
Come join the OSU proteomics community! We're hiring a senior staff scientist at @OhioStatePIIO run by @Zihai to direct a mass spec proteomics facility to monitor immune proteins in cancer. A great opportunity for independent MS researchers! DM for details osu.wd1.myworkdayjobs.com/OSUCareers/job…
Great seeing this big collaborative effort come together with @lea_starita, @Nobu_Hamazaki, @JShendure, and led by @riddhimankg! A rich resource to understand mammalian gastrulation using organoid models, with lots to explore at: gastruloid.brotmanbaty.org biorxiv.org/content/10.110…
Very happy to share our latest w/ @dschweppe1 and @Nobu_Hamazaki labs, where we map the temporal dynamics and proteomic landscape of mammalian gastruloids. Sneek Peak 🧵below #stembryo #gastruloid #proteomics biorxiv.org/content/10.110…
This should be an exciting, educational lecture on ion traps! I am really looking forward to moderating audience questions for Philip during his talk tomorrow. Come join us!
Register today for this powerful webinar organized by HUPO’s ETC – “The Past, Present and Future of Quadrupole Ion Trap Mass Spectrometers in Proteomics” Speaker Philip M. Remes, PhD – Thurs. Sept. 12 at 9 am PDT, 12 pm EDT, 5 pm CET, 12 am CST us06web.zoom.us/webinar/regist…
Kudos to the team of @briansearle, @ArianaShannon, et al. Using a linear ion trap with 1ng total protein on column, they found clear consistency of #ImmuneMarkers expression between #ImmuneCell populations measured using #FlowCytometry and #proteomics go.osu.edu/CrVb
Come see the Searle lab at ASMS Anaheim #asms2024
(BioRxiv All) Hybrid Quadrupole Mass Filter Radial Ejection Linear Ion Trap and Intelligent Data Acquisition Enable Highly Multiplex Targeted Proteomics dlvr.it/T7jjN6 (RSS) #BioRxiv #MassSpecRSS