
Ava Amini
@avapamini
principal researcher @MSFTResearch | AI for biomedicine | instructor @MITDeepLearning | alumna @MIT @Harvard
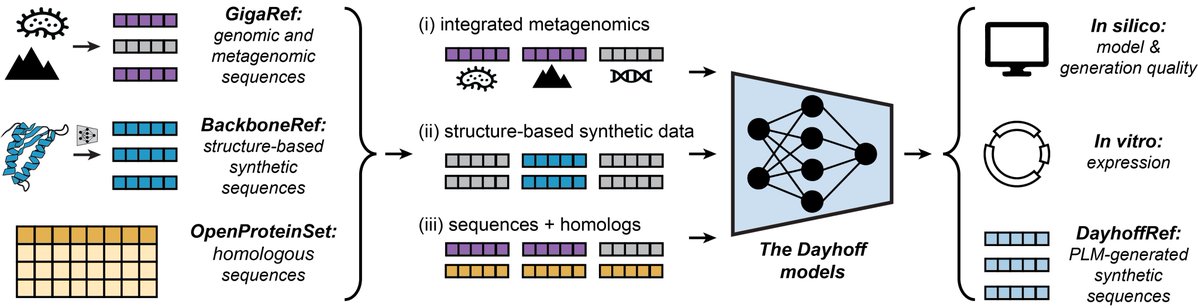
thrilled to share The Dayhoff Atlas of protein language data and models 🚀 protein biology in the age of AI! aka.ms/dayhoff/prepri… we built + open source the largest natural protein dataset, w/ 3.3 billion seqs & a first-in-class dataset of structure-based synthetic proteins
The Dayhoff Atlas! Open code. Open weights. Open datasets. Thanks @huggingface for helping to facilitate open science. huggingface.co/collections/mi… @ClementDelangue @julien_c
Our models, code, and data are openly available on Github, Zenodo, and Huggingface. huggingface.co/collections/mi… zenodo.org/records/152652… github.com/microsoft/dayh…
**A grand unified theory on what will happen in biotech in the next 10-20 years** the two major forces reshaping industrial biotech in the next decade are: 1. China 2. AI - and they're critically linked how? China's low R&D cost basis democratizes execution by providing…
New synthetic and metagenomic data boosted experimental success while popular metrics failed to predict it. Read Ava's thread on the really cool models, analysis, and data resources!
increasing model and data scale increased the fraction of proteins expressed by E. coli, and the highest expression success rate came from augmenting w/ structure-based synthetic data. data quality + diversity bring real gains in real-world protein expression!
🧬 The largest open dataset of natural proteins in the world — 3.3 billion seqs 🧠 A 3 billion param hybrid ssm+transformer model 🤗 Fully open-source data + model biorxiv.org/content/10.110… Congrats to @avapamini + entire team, including @LiquidAI_'s own Kaeli Kaymak-Loveless
thrilled to share The Dayhoff Atlas of protein language data and models 🚀 protein biology in the age of AI! aka.ms/dayhoff/prepri… we built + open source the largest natural protein dataset, w/ 3.3 billion seqs & a first-in-class dataset of structure-based synthetic proteins
In 1965, Margaret Dayhoff published the Atlas of Protein Sequence and Structure, which collated the 65 proteins whose amino acid sequences were then known. Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
Why is this not all over my feed all day today?!?! 😁@avapamini @KevinKaichuang The Dayhoff Atlas: scaling sequence diversity for improved protein generation biorxiv.org/content/10.110…
I was surprised to see that BackboneRef boosts Dayhoff‑170 m pLM generations expressed in E. coli 27.6% → 51.7%, 1.9× with zero filtering ...while common metrics (pLDDT, perplexity) failed to predict wet‑lab outcomes (AUROC ≤ 0.57) This quietly re‑prioritizes how we…
👀#DayhoffAtlas dropped for #SynBio:👀 3.34B natural🧬 + 46M structure‑guided synthetic protein sequences (from 240k novel backbones), all open‑source Hybrid Mamba‑Transformer learns single seqs & MSAs → 51.7 % of unfiltered designs express in E. coli🦠✨…
To the GPU-poor grad students out there, finding a better predictor of expression is one of the highest leverage contributions you could make to PLM research. Scale isn't always all you need.
I was surprised to see that BackboneRef boosts Dayhoff‑170 m pLM generations expressed in E. coli 27.6% → 51.7%, 1.9× with zero filtering ...while common metrics (pLDDT, perplexity) failed to predict wet‑lab outcomes (AUROC ≤ 0.57) This quietly re‑prioritizes how we…
Data diversity, quality & relevance rules over model size any day of the week. Very clever approach of generating synthetic protein sequences from backbone structures to give big boosts to pLMs.
Learning on GigaRef yielded a small increase in the fraction of expressed proteins. Increasing model and dataset scale further improved the expression rate. Augmenting training with structure-based synthetic data from BackboneRef produced the highest expression success rate.
Very cool work on scaling data for protein language modeling, congrats to the team!
In 1965, Margaret Dayhoff published the Atlas of Protein Sequence and Structure, which collated the 65 proteins whose amino acid sequences were then known. Inspired by that Atlas, today we are releasing the Dayhoff Atlas of protein sequence data and protein language models.
Anybody can run cloud LLMs — that's the past. Now with LEAP 🐸, you don’t need the cloud — just tap. No lag, no limits, no looking back.
Today, we release LEAP, our new developer platform for building with on-device AI — and Apollo, a lightweight iOS application for vibe checking small language models directly on your phone. With LEAP and Apollo, AI isn’t tied to the cloud anymore. Run it locally when you want,…
🚀 Introducing LFM2: the fastest on-device foundation models on the market. Built by @LiquidAI_, LFM2 is: ⚡️ Optimized for speed, quality, efficiency. No trade-offs. 🤗 Available in three sizes: - LFM2-350M - LFM2-700M - LFM2-1.2B 🧠 SoTA on knowledge, math, and…
Today, we release the 2nd generation of our Liquid foundation models, LFM2. LFM2 set the bar for quality, speed, and memory efficiency in on-device AI. Built for edge devices like phones, laptops, AI PCs, cars, wearables, satellites, and robots, LFM2 delivers the fastest…