Chris Parker
@_chrisgparker_
Professor @scrippsresearch | Department of Chemistry | chemical biology & chemical proteomics
Check out our recent Opinion in @TrendsinPharma on chemical proteomic binding site determination for non-covalent compounds
Chemical proteomic mapping of reversible small molecule binding sites in native systems dlvr.it/TFMRnc
We are excited to kick off our series of 4 manuscripts from the RESOLUTE consortium by sharing the metabolic mapping of the Solute Carrier Superfamily (biorxiv.org/content/10.110…)!
A big congratulations to the newly minted Dr. Forrest @InesForrest . Super proud!
Proud to share that I have officially completed my PhD and graduated from @scrippsresearch! 🥳 Grateful to my mentors, collaborators, friends, and family for their support throughout this challenging yet rewarding and unforgettable journey! 🧬 Excited for the next chapter! 🎓
I’m a standing member on the Chemical Biology and Probes (CBP) NIH study section that was supposed to meet tomorrow. It just got postponed this afternoon with a date yet to be determined….
Check out our recent efforts in @ChemRxiv to expand the chemical tractability of the proteome through NP-inspired photoaffinity probes. chemrxiv.org/engage/chemrxi…
Congratulations to Scripps Research Prof. Benjamin Cravatt, PhD, on receiving the 2025 William H. Nichols Medal from @AmerChemSociety for his pioneering work in activity-based protein profiling, advancing drug discovery for cancer & neurological disorders. ow.ly/pbwV50UBnm9
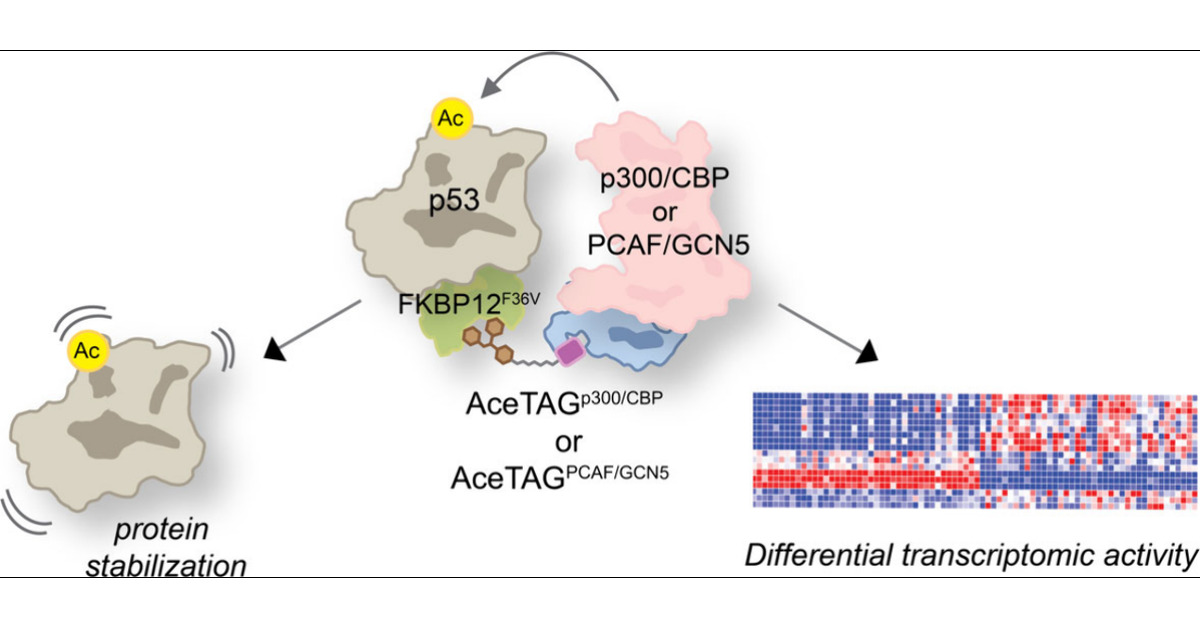
Great paper by the group of @_chrisgparker_. They further developed their AceTAG technology to also recruit the lysine acetyltransferase PCAF/GCN5. Together with their p300/CBP recruiter, this allowed in-depth insights into the function of p53 acetylation pubs.acs.org/doi/full/10.10…
the first project of my PhD is now out @J_A_C_S ! super proud to introduce ABAP (my first acronym!) and share our findings on how lysine acetylation modulates protein function & interactions. so surreal to see this in print pubs.acs.org/doi/10.1021/ja…
Proud to share my first, first-author paper of graduate school! This year-long detour started with “we made a neat tool - what biology can we figure out with it?” Turns out we can find regulators of placental cell differentiation. pubs.acs.org/doi/10.1021/ac…
Excited to share a key part of my postdoctoral work in the Cravatt lab at @ScrippsResearch, published today in @NatureChemistry! Huge thanks to collaborators @brumelillo, @_RachelHayward, @VividionRx, and all coauthors.
Excited to share this @CellChemBiol Review I wrote with Ben Cravatt highlighting recent applications of ABPP for ligand discovery. We highlight the expansion of ABPP beyond enzyme active sites with a special focus on covalent ligands. Enjoy! tinyurl.com/48cyaj6d
Very honored to be one of 19 graduate students recognized by the @AmerChemSociety and @ACSGradsPostdoc for leadership in the important mission of promoting diversity, equity, inclusion and respect! Congrats to all the other awardees, together we are making a difference! 🌟
Happy to share new work led by recent grad @AnnyChe77788549 and @SinghaSoumya. Here, we expanded our AceTAG platform to include PCAF/GCN5 & used this toolbox to investigate functional consequences of targeted p53 acetylation. w/@Michael_A_Erb. @ACSBioMed pubs.acs.org/doi/full/10.10…
pubs.rsc.org/en/content/art… No 1⃣1⃣ Check out this intriguing mechanistic study on dialkyldiazirines by @FoxGroupUD (@UDelaware), @_chrisgparker_ (@scrippsresearch), @ChrisamEnde1 (@pfizer). Grateful to be part of this work all the way from @univienna!! @azidoalcohol @MaulideLab
Congrats to Benjamin Cravatt, PhD, the Norton B. Gilula Chair in Biology and Chemistry at Scripps Research, on receiving the 2024 Heinrich Wieland Prize. His work on disease-causing proteins and activity-based protein profiling is advancing drug discovery. ow.ly/Jgze50T4xUE
I’m incredibly honored to join @HHMINEWS community. Deeply grateful to my lab, colleagues, collaborators, and mentors. I’m lucky to have you all.
⭐️ Big news! HHMI is investing $300+ million in 26 new #HHMIInvestigators from 19 US institutions. These top scientists will drive groundbreaking research in diverse fields, from neuroscience to immunology to structural biology & beyond. Welcome to HHMI! hhmi.news/4df2eVs
Big day for the lab as our MultiMap platform is now published on Science! Congratulations to Lindsey @Zhi_Lindsey_Lin and the team! science.org/doi/10.1126/sc…
Thrilled to share our MultiMap paper is now published online on Science. @ScienceMagazine With @realJimWells, we present a multiscale photocatalytic proximity labeling platform for interactome profiling. science.org/doi/10.1126/sc…
8 new colleagues are joining the @scrippsresearch faculty! This includes current @HHMINEWS investigators Doro Kern & Darrell Irvine, Meng Zhang from Xiaowei Zhuang's lab, and Ann Kennedy @Antihebbiann in Neuroscience. Exciting times ahead! 🚀 scripps.edu/news-and-event…