Torgeir R Hvidsten
@TRHvidsten
Bioinformatics, Gene regulatory networks, Comparative regulomics; in plants, salmonids and more. Professor @UniNMBU
It is still possible to apply for PhD scholarships within computational host/microbiome genomics (hologenomics) at the Norwegian University of Life Sciences! The positions are part @HoloGen_MSCA jobbnorge.no/en/available-j… Deadline: January 17th. @SabinaLLaRosa @ThePopeLab
🥳I'm incredibly honoured to receive the @MicrobioSoc Peter Wildy Prize 2025 for #scicomm, a true team prize! Huge thanks to those who nominated me & to incredible collaborators & team members. So many people to thank - I'll be highlighting them all in my prize lecture!🦠
Peter Wildy Prize 2025: Professor Lindsay Hall (@hall_lab), University of Birmingham, UK (@unibirmingham)
A multi-omic approach enables the reconstruction of microbial metabolic dynamics in the salmon gut in response to feed and feed supplements 🐟@SabinaLLaRosa @TRHvidsten @ThePopeLab @shashankguptapn doi.org/10.1038/s42003…
We are seeking three PhD students within computational biology/bioinformatics at the Norwegian University of Life Sciences: Comparative evolutionary genomics: jobbnorge.no/en/available-j… Host-microbe interactions using genomics: jobbnorge.no/en/available-j… Deadlines: October 31st.
Are you interested in learning about microbes living in the salmon gut? Then check our new paper by @SabinaLLaRosa @kid4_j @srsandve @MortenLimborg and @ThePopeLab nature.com/articles/s4156…
4 years of collaboration has poured into this resource! Massive thanks to @SabinaLLaRosa and @quark_buzo_vera for driving this and of course all our partners coming from @UniNMBU and abroad @srsandve @TRHvidsten @MortenLimborg @ines_thiele @Stebe69
A fishy tale about the the Salmon Microbial Genome Atlas is now out in @NatureMicrobiol. We hope this culture and genome biobank provides a functional resource to the community and supports the next generation of salmon gut microbiome projects. nature.com/articles/s4156…
We are seeking a PhD student within comparative evolutionary genomics. What happens with gene regulation after the genome is doubled? Help us find out using RNA-seq, ATAC-seq and Hi-C data! Supervisors: @TRHvidsten and @srsandve jobbnorge.no/ledige-stillin…
Another exciting #phdpositions advert up for our @HoloGen_MSCA Doctoral network! Check out the details below 😃👇
📢 We are hiring a fully salaried #PhDstudent in #holobiomics of Resource Efficiency of Atlantic salmon🧬🐟 @UniNMBU 🇳🇴 @srsandve #microbiability The position is part of the HoloGen MSCA Doctoral Network. @HoloGen_MSCA Please RT. 📅 deadline 25/10/24 jobbnorge.no/en/available-j…
We are looking for two PhD students to join our @UniNMBU team to explore host-microbe interactions using genomics. The positions are part of HoloGen, a MSCA Doctoral Network. @HoloGen_MSCA @ThePopeLab @SabinaLLaRosa Apply here: jobbnorge.no/en/available-j…
Open PhD position in computational microbiome research, covering population cohorts, microbial ecology, epidemiology & multi-omic integration. In HoloGen EU doctoral network / international applicants are eligible. Turku, Finland. PM for details.
We have an open position! We are seeking a doctoral candidate who will do #metabolomics and multiomics research related to #microbiota-host interactions in the @MSCActions doctoral network @HoloGen_MSCA 3 year fully funded position! afekta.com/news/open-posi…
Check out these exciting @HoloGen_MSCA #PhDposition's @UniTurku! 👇💻🧬💩 ats.talentadore.com/apply/1-2-full… Please also note @MSCActions PhD's have a mobility rule ➡️can't have lived/worked in host country for >12 months within the 3 years before Sept 2024.
Super excited to be part of the new @MSCActions doctoral network @HoloGen_MSCA that will train the next generation of researchers in hologenomics! Look out for positions in the coming weeks.
🚀 Exciting news! The new @MSCActions Doctoral Network @HoloGen_MSCA is kicking off on Sept 9th! Led by Prof. @TRHvidsten @UniNMBU with 8 EU institutions, we’re diving into the cutting-edge field of *hologenomics* to uncover how microbes and host genetics interact. 🌍🧬#HoloGen
In a perfect microbial world we would all work with genomes/MAGs/isolates…but we are not there yet, and the old workhorse 16S rDNA gene still has a role to play. If you missed @oveoyas poster at #ISME19 check out the preprint biorxiv.org/content/10.110…
Check out our new review on promising methods for analyzing omics data from matching host and microbiome samples (holo-omics)! pubs.rsc.org/en/content/art… with: @kobelkobelkobel @IdunBurgos @oveoyas @ThePopeLab @velma_te_a

Congratulations!!! This is the worst kept secret in all of ML.
How did these authors miss the opportunity to paraphrase Watson and Crick? “It has not escaped our notice that these findings have implications for the capabilities of large language models/AI”. nature.com/articles/s4158…

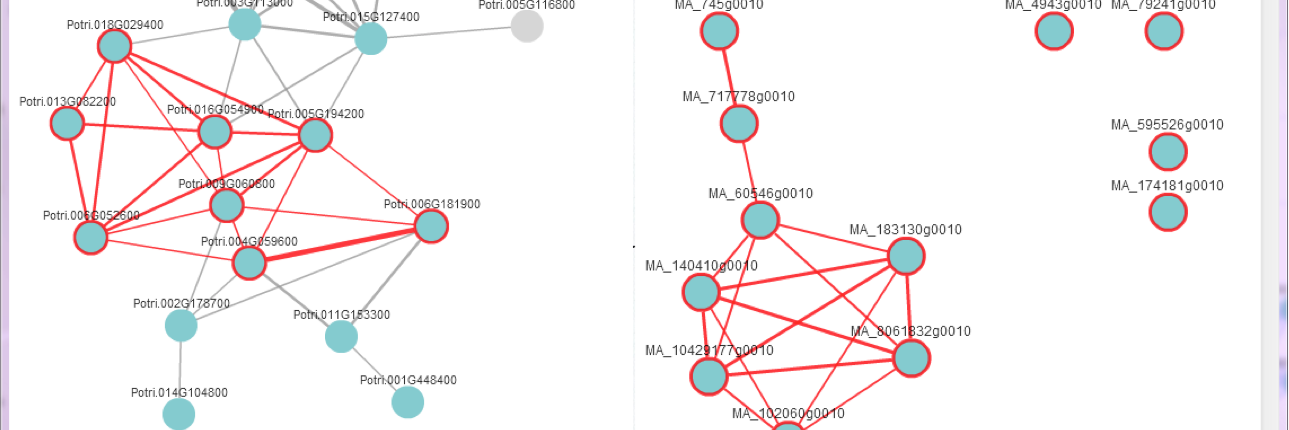
Delighted to see that our Populus super-pangenome study is featured as the cover story in the latest issue of Molecular Plant!
Oh nice! @Nature publishes another amazing advertisement for an amazing product masquerading as a scientific publication with no code, no model & a webserver that allows 10 queries at a time from a billion dollar company. But it's "gold" open access folks! Rejoice!