
Ronald Breaker
@RonBreaker
Prof. Breaker is an RNA biochemist, HHMI Investigator, Sterling Professor, and Chair of Molecular Biophysics and Biochemistry at Yale University, New Haven, CT.
Computational Biophysics and Biochemistry: Faculty Position Opening in the Department of Biophysics and Biochemistry at Yale.
My Yale team has exciting RNA research advances related to neurological disorders, with relevance to #bipolar, #autism, #Parkinson’s, and more. Seeking private or commercial funding for our next steps. Possible partners DM @RonBreaker or contact [email protected] for info.

In the new year, I will try to post the most exciting Breaker Lab science content on my #Bluesky account at... @ronbreaker.bsky.social More interesting #riboswitch finds and other nucleic acid oddities are likely to come!
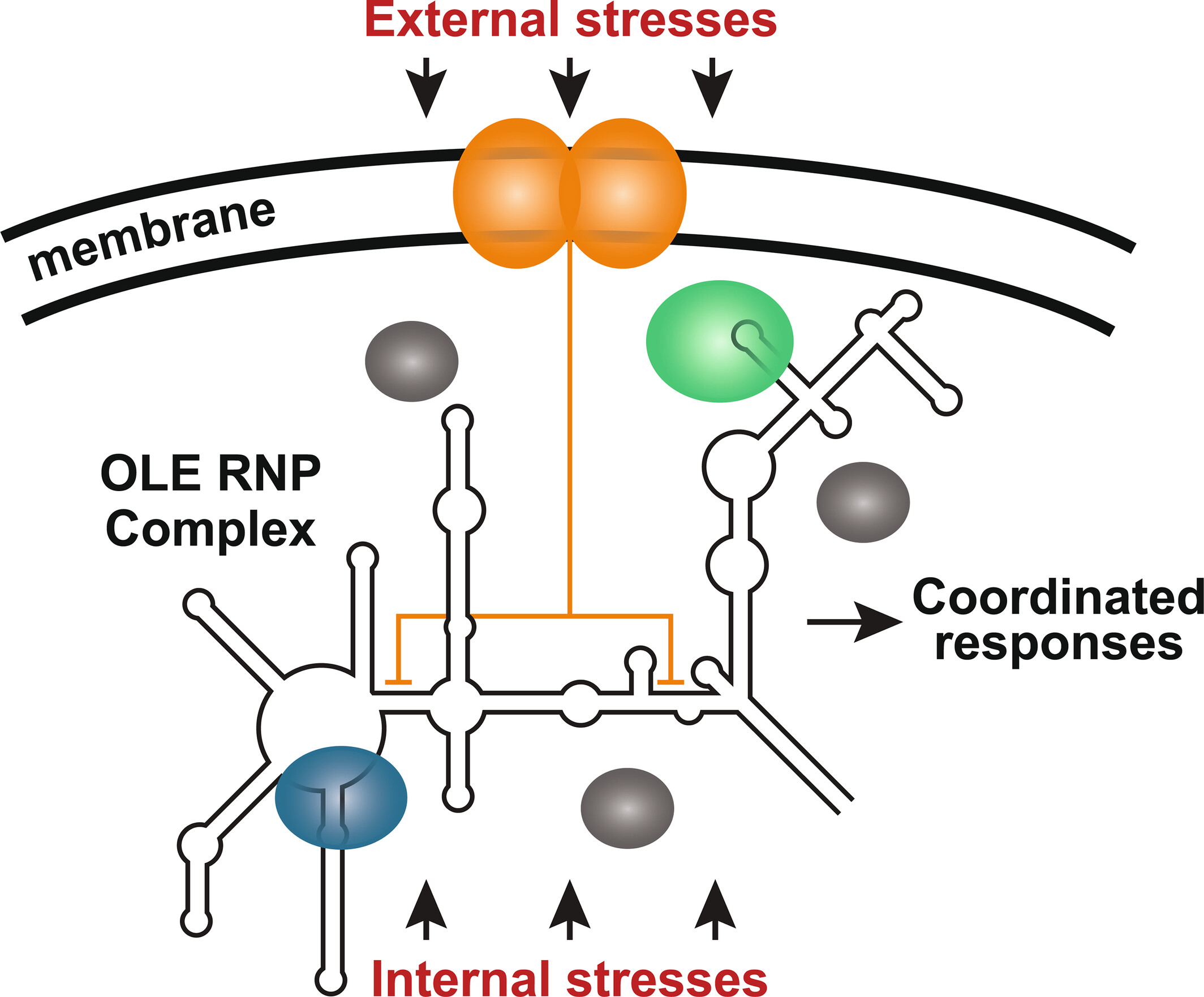
#ncRNA molecules are stranger than you think. We propose that OLE RNA is bacteria's functional mimic of #mTOR. The Das lab at Stanford proposes OLE RNA penetrates cell membranes! They also find that RNAs we called ROOL and GOLLD form 8-membered nanocages. biorxiv.org/content/10.110…
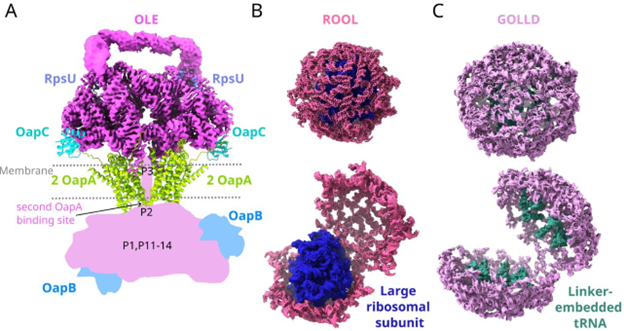
OLE #noncoding RNA always partners with OapA protein (unknown function). Chrishan Fernando used a computational approach to find that OapA anticorrelates with MpfA - a proven metal ion transporter. See @thechrishan for his posts on this odd #ncRNA story. tinyurl.com/yt48x8pr

Here is a recent posting for a Lecturer position with the Department of Molecular Biophysics and Biochemistry (MB&B) at #Yale University. Please consider joining our research and educational community @YaleMBB and contributing to these important missions! apply.interfolio.com/145019

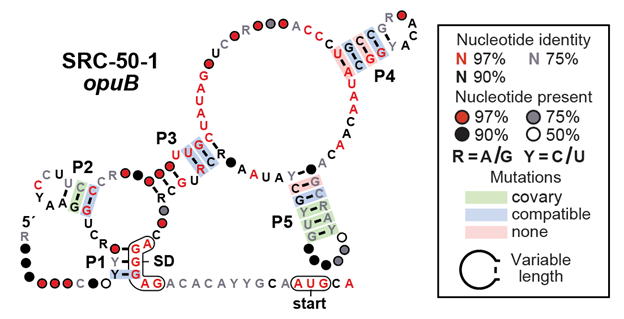
1000s of ligand-binding #riboswitch classes likely remain hidden in bacterial transcripts. In @NAR_Open, we report many new candidate #ncRNA classes (including the opuB riboswitch candidate shown below) discovered by careful bioinfo analysis of 50 genomes. tinyurl.com/yj2n47sj
X-ray structure of the azaaromatic #riboswitch in @CellDiscovery by Joe Piccirilli and colleagues @UChicago reveals how a #ncRNA forms a pocket that broadly binds planar, hydrophobic ligands. A parallel-eye stereoview shows plenty of base-ligand stacking. nature.com/articles/s4142…

The bacterial OLE #noncoding RNA is needed for cells to grow on most carbon/energy sources (except glucose or citrate) and to secrete proteins. This find suggests OLE (#ribozyme?) is part of a major regulatory apparatus of many Gram+ species. @PNASNexus tinyurl.com/5bf45s2h

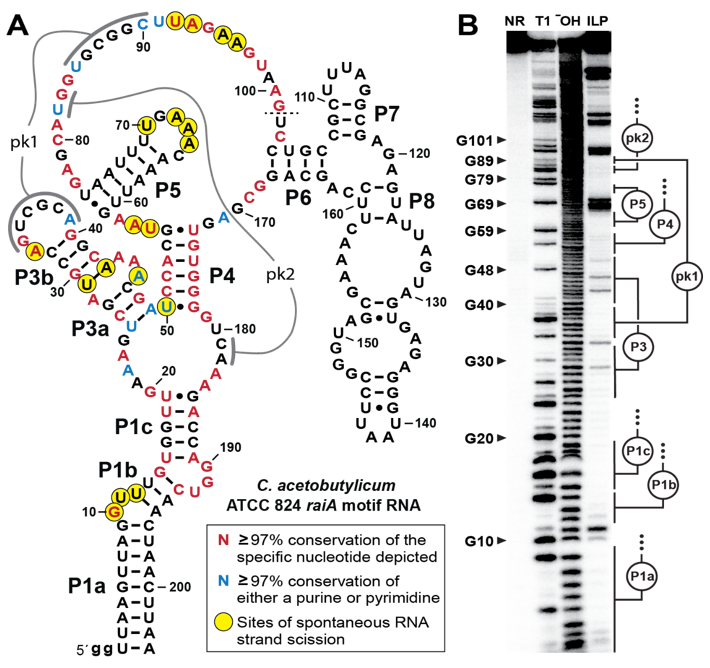
Large, conserved #noncoding RNAs in bacteria with major biochemical functions are rare. Here is #raiA RNA, which has the highest % of strictly conserved nts of any #ncRNA known, and a complex structure. This must be a #ribozyme. See article at tinyurl.com/2h7heypd. @PNASNews
Yale MB&B is seeking a Lecturer to contribute to our educational mission. Please consider joining our scientific community and using your skills for teaching biochemistry. You will work with fantastic colleagues and enthusiastic students! @YaleMBB apply.interfolio.com/114946

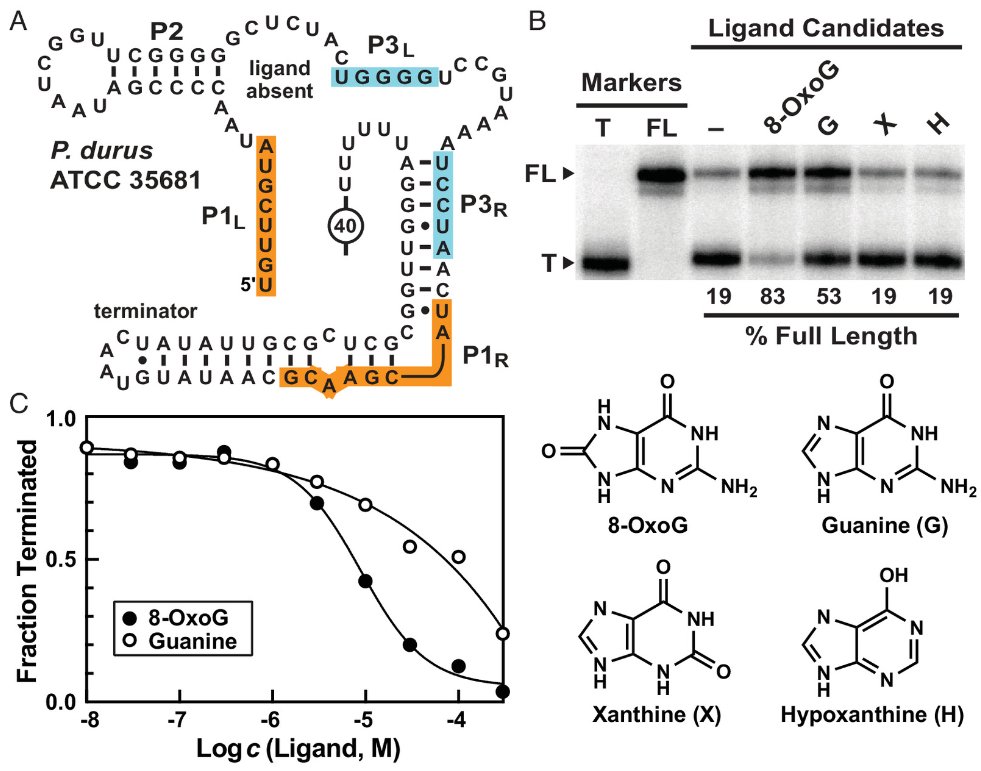
The discovery of a #riboswitch class for 8-OxoG expands the diversity of #purine bases sensed by #aptamer RNAs. The image depicts sites of purine modification (colored boxes) relevant to the riboswitch classes whose names are in bold. @PNASNews #RNAWorld pnas.org/doi/10.1073/pn…
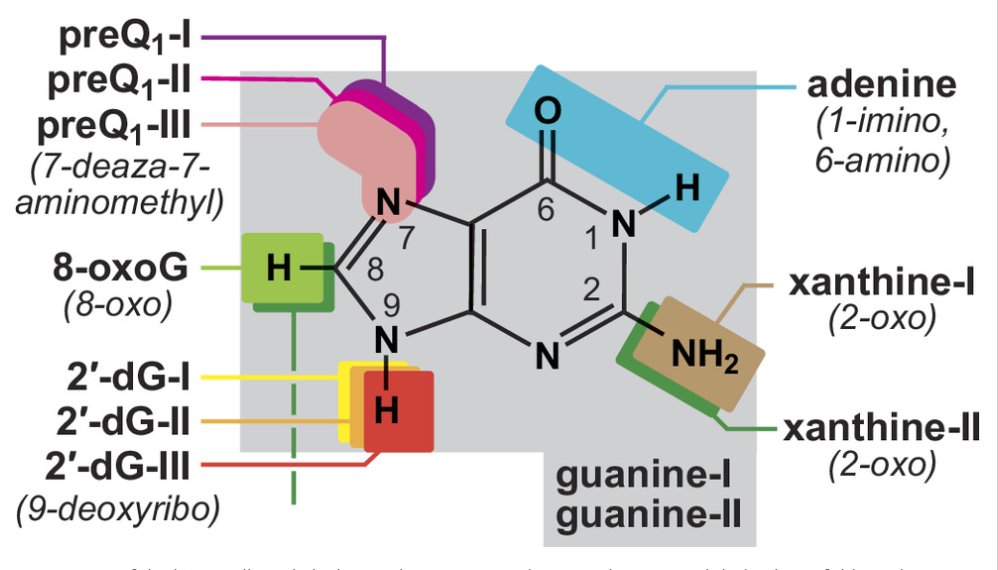
A variant of #guanine #riboswitch RNAs has adapted to sense the modified base 8-oxoguanine (8-OxoG). Oxidizing agents that damage DNA, such as hydrogen peroxide, activate gene expression via this #ncRNA device. @PNASNews pnas.org/doi/10.1073/pn…
Bacteria’s Brain? The mysterious OLE #ncRNA appears to be bacteria’s functional equivalent of eukaryotic #mTOR. The #OLERNA component of the #bTOR (bacterial Tasks OLE Regulates) complex looks like an #RNAWorld relic. @MolMicroEditors doi.org/10.1111/mmi.15…
Variant #ppGpp #riboswitch class RNAs found by Jonathan Jagodnik, Rick Gourse and colleagues @UWMadison to have tuned their specificities to favor ppGpp, #pppGpp, and maybe other biomolecules. Findings showcase RNA functional diversity. @JonathanJagSci pubmed.ncbi.nlm.nih.gov/36617997/

In @NAR_Open, with postdoc @mgmo7sen, undergrad Matt Midy, and grad student @Aparaajita_B, we exploit the guanine-I #riboswitch architecture to engineer genetic switches for #quinine and #caffeine. Next challenge: switches for gene therapy applications. doi.org/10.1093/nar/gk…

X-ray model of an NAD+-II #riboswitch by the Micura and Ren labs in @NAR_Open reveals how #ncRNA senses the redox state of a ubiquitous enzyme cofactor. The #aptamer stacks G6 and G42 on the plus-charged nicotinamide ring (cation-pi interaction), and rejects NAD's puckered ring.

#Yale undergrad Matt Midy (center) representing the Breaker lab at #ABRCMS2022 by winning a poster award on his work to create synthetic #Riboswitch RNAs for #Quinine and #Caffeine. Paper under review. Grad School admission committees: watch for his application next year! @ABRCMS
Another update on the mysterious OLE #ncRNA from bacteria. In @jbiolchem we report a third essential protein partner called OapC, which alters RNA folding to form a k-turn. We still think OLE is a #ribozyme relic from the #RNAWorld. pubmed.ncbi.nlm.nih.gov/36336078/

Two bacterial #Riboswitch classes sense #lithium ions, activating expression of pumps that eject this toxic ion. Our paper in @SciReports reveals how these small #ncRNA domains fold, activate genes (see photo), and sometimes mutate to sense #sodium ions. nature.com/articles/s4159…
