
Oxford Protein Informatics Group (OPIG)
@OPIGlets
Research group led by Charlotte Deane, based in the Department of Statistics at the University of Oxford.
Was an honour for our collaboration with Sarosh Irani's group to be recognised with the PNAS Cozzarelli Prize #immunoinformatics #encephalitis #autoreactivity #bcells
Join us in celebrating our 2024 Cozzarelli Prize Class IV: Biomedical Sciences winning paper, “Ultrahigh frequencies of peripherally matured LGI1- and CASPR2-reactive B cells characterize the cerebrospinal fluid in autoimmune encephalitis.” Read more: ow.ly/SyAL50VyXLV
AI is changing the discipline of Biology- but can it crack one of science’s hardest problems: drug discovery? At the center of that challenge is Professor Charlotte Deane, who joins us at #ISMBECCB2025! @iscb @OPIGlets youtu.be/PyZbeoLsLuQ
Earlier this week at ISMB, Prof. Charlotte Deane spoke to the @WebsEdgeTV team for @iscb Spotlight TV. Check out what she had to say about AI, biological datasets, drug discovery, and so much more youtu.be/PyZbeoLsLuQ?si… instagram.com/p/DMaQVr8tEFj/…
Our work exploring the ability of and requirements for ML to predict the effects of mutations on antibody–antigen binding affinity (ΔΔG) is out now in @NatComputSci!
Out now! @AlissaHummer, @OPIGlets and colleagues present Graphinity, a method to predict change in antibody-antigen binding affinity (∆∆G). Featuring synthetic datasets of ~1 million FoldX-generated and >20,000 Rosetta Flex ddG-generated ∆∆G values! nature.com/articles/s4358…
Out now! @AlissaHummer, @OPIGlets and colleagues present Graphinity, a method to predict change in antibody-antigen binding affinity (∆∆G). Featuring synthetic datasets of ~1 million FoldX-generated and >20,000 Rosetta Flex ddG-generated ∆∆G values! nature.com/articles/s4358…
Professor Charlotte Deane is speaking this Thursday, 3rd July at her former college (Univ) as part of a session on Creativity and AI. Online and in-person tickets available (free for students) Profile: univ.ox.ac.uk/news/profile-c… More details here: univ.ox.ac.uk/event/univ-sem…

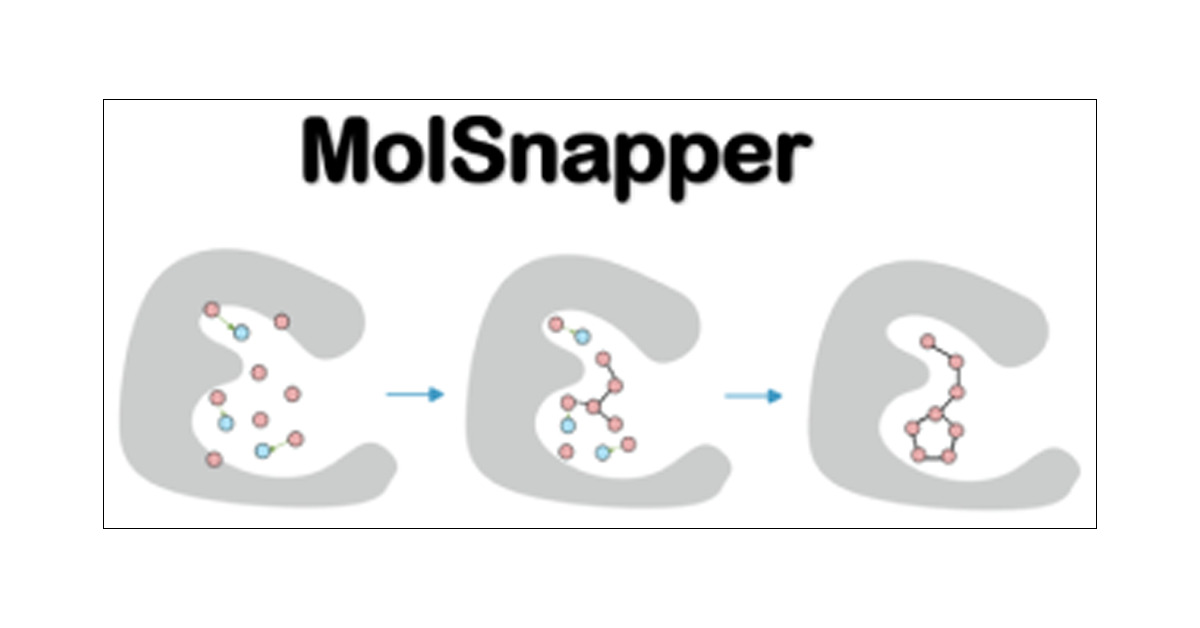
MolSnapper: Conditioning Diffusion for Structure-Based Drug Design #DrugDesign pubs.acs.org/doi/10.1021/ac… @YaeliZiv @fergus_imrie @bmarsden19 @OPIGlets #JCIM Vol65 Issue9 #MachineLearning #DeepLearning
Come and find OPIG at #PEGSummit today (Thursday) C089: LICHEN: Light-Chain Immunoglobulin Sequence Generation Conditioned on the Heavy Chain and Experimental Needs - Henriette Capel C090: Predicting the Developability of Nanobodies to Improve Therapeutic Design - Gemma Gordon
🚀 LLMs can now do chemistry! Our new preprint shows that state-of-the-art reasoning models can now perform advanced chemical reasoning tasks, without any assistance from external tools. Here’s what o3-mini can already do👇 (1/🧵)
Do you wish working with T-cell receptor structures was easier? Us too! STCRpy, our software suite for TCR structure parsing, interaction profiling and machine learning dataset preparation is now available! Github: github.com/npqst/stcrpy/ Pre-print: doi.org/10.1101/2025.0… 1/3
MolSnapper has now been published in @JCIM_JCTC! MolSnapper integrates expert knowledge into diffusion models for structure-based drug design using a conditioning approach. Congratulations @YaeliZiv, @fergus_imrie, Brian Marsden, and Charlotte Deane. pubs.acs.org/doi/10.1021/ac…
3-year postdoc opportunity as part of the Novo Nordisk - Oxford Fellowship programme! Develop machine learning approaches for drug discovery with me, Charlotte Deane (Oxford), and Christos Nicolaou (Novo Nordisk). 1 week left to apply! Details in next post