Kurumizaka Lab
@KurumizakaLab
The University of Tokyo / Chromatin / nucleosome/epigenetics/The singing structural biologist
Check out our review article on noncanonical nucleosomes, which differ from canonical nucleosomes in their histone composition and DNA wrapping. The article highlights recent insights into their structural and functional diversity. sciencedirect.com/science/articl…
With collaboration with Sugiyama lab(Kyoto Univ.), Our work regarding Cryo-EM and SAXS analysis of overlapping dinucleosome has been published! doi.org/10.1093/pnasne…

Our laboratory website is now available in English! Check out our research! iqb.u-tokyo.ac.jp/kurumizakalab/…
Our new paper has been published! In this study, we revealed the mechanism by which the efficient transcription by RNA polymerase II is accomplished on nucleosomes containing a histone variant H2A.B. #cryoEM embopress.org/doi/full/10.10…
Check out our new preprint on H3-H4 octasome transcription by RNAPII! We discovered that H3-H4 octasome is transcribed more efficiently than nucleosomes and visualized its transcription process by cryo-EM. #cryoEM doi.org/10.1101/2025.0…
Check out our methods paper on high-resolution cryo-EM analyses of nucleosomes is published! It covers nucleosome preparation, grid making, and single-particle analysis for cryo-EM. This work is a collaboration with the Danev Lab at the University of Tokyo.link.springer.com/protocol/10.10…
Check out our paper on the cryo-EM analysis of chromatin derived from human cells! We have established a technically accessible method for analyzing the chromatin structure prepared from human cells. onlinelibrary.wiley.com/doi/10.1111/gt…

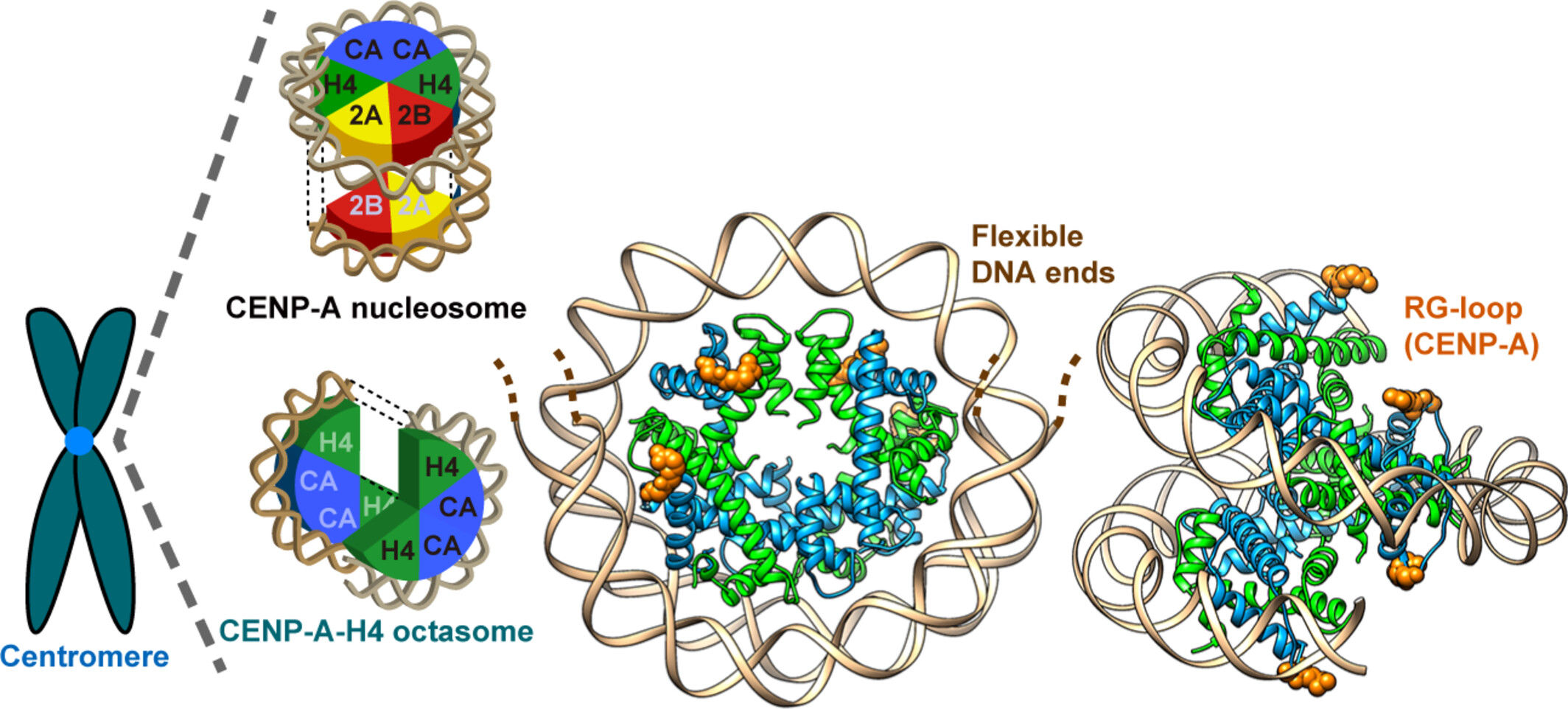
Check out the cryo-EM structure of the CENP-A-H4 octasome. This nucleosome-like structure contains a histone octamer composed solely of centromere specific histone variant, CENP-A, and H4! Amazing work by @NozawaL! Congratulations! 🎉 #Subnucleosome onlinelibrary.wiley.com/doi/10.1111/gt…
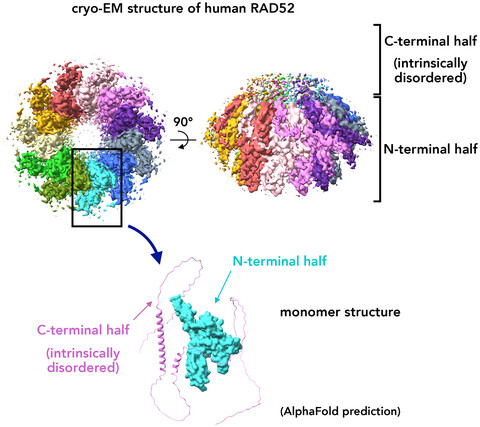
Our paper, “The cryo-EM structure of full-length RAD52 protein contains an undecameric ring,” is among the top 10 most-cited papers in FEBS Open Bio (2023)! Great collaboration with Kagawa-lab (Meisei Univ.)—congrats, Kagawa-group! 🔗 doi.org/10.1002/2211-5… #CryoEM #RAD52
Excited to share our paper on the RNAPII-NELF-DSIF-TFIIS-nucleosome complex published @ScienceAdvances , in collaboration with the Sekine lab @RIKEN_BDR ! This study reveals how NELF and nucleosome function in promoter-proximal pausing of RNAPII! doi.org/10.1126/sciadv…
Check out our paper on the chromatin protein DEK published @NatureSMB! We found DEK as a nucleosome binding protein and its role in promoting H3K27me3 by PRC2! Great collaboration with Gotoh Lab, @YusukeKishi1 , MasumotoG, and @Cell_Tokyo_Tech ! Thank you! nature.com/articles/s4159…
Super excited to share our work on the chromatin-associated oncoprotein DEK published in @NatureSMB!!😆 We found DEK as a nucleosome binding protein that facilitates H3K27me3 by PRC2 through chromatin reorganization!! Check out! rdcu.be/eaSK6
Our new paper about the cryo-EM structures of barrier-to-autointegration factor (BAF) and Lamin A/C complexed with nucleosomes has been published! This study reveals that BAF and Lamin A/C interact with nucleosomes in multiple binding modes. rdcu.be/d9jmd
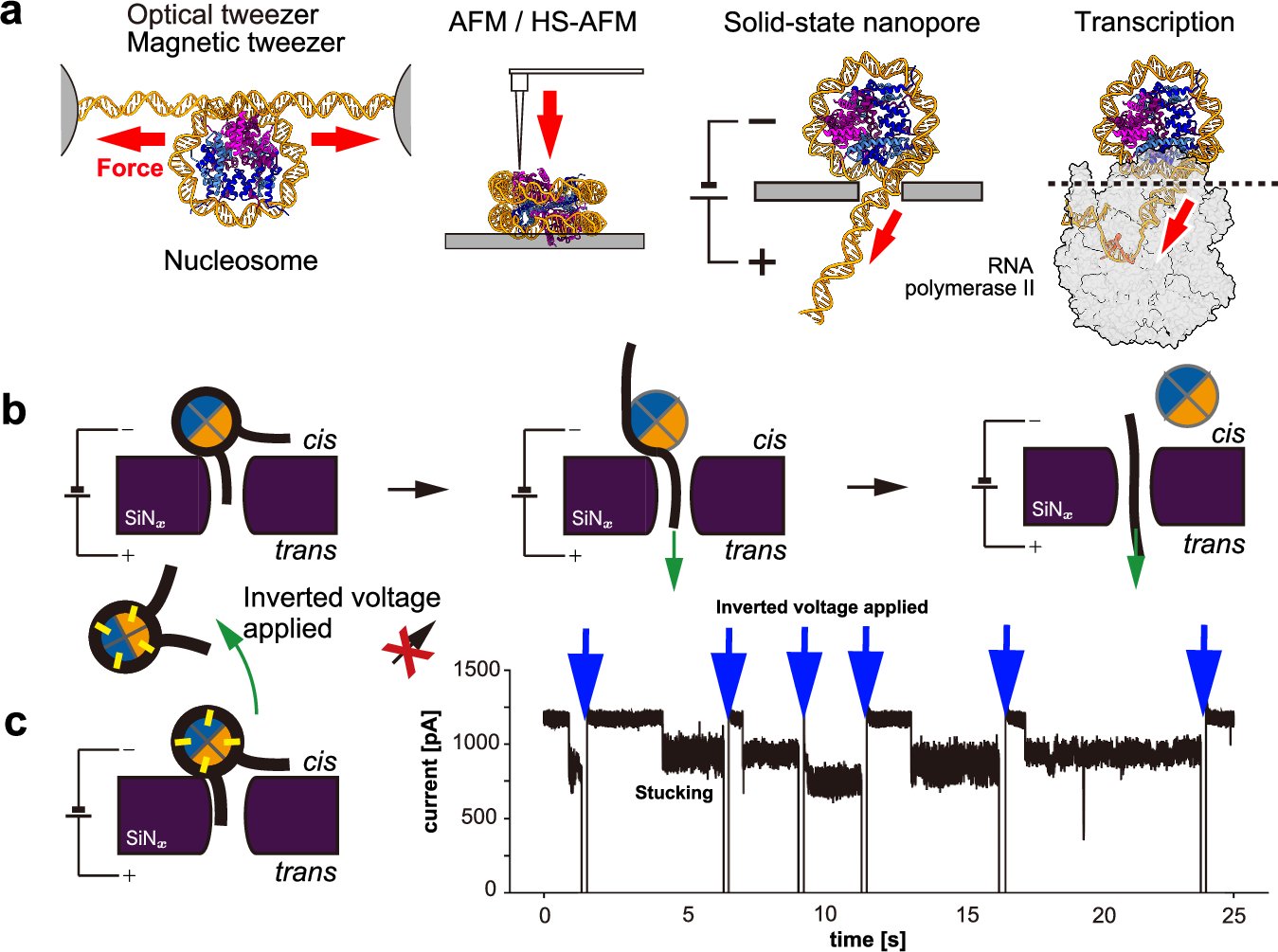
A new paper has been published! Congratulations to the Uemura lab (Univ of Tokyo) and Takada lab (Kyoto Univ) for our collaborative work on observing nucleosome unwinding using solid-state nanopores! nature.com/articles/s4200…
Very excited to share that our new work is finally out🎉 We demonstrate that the chromatin remodeling factor DDM1 “opens” the nucleosome structure. nature.com/articles/s4146… You can also access to the PDF file as bellow: rdcu.be/dNq8u
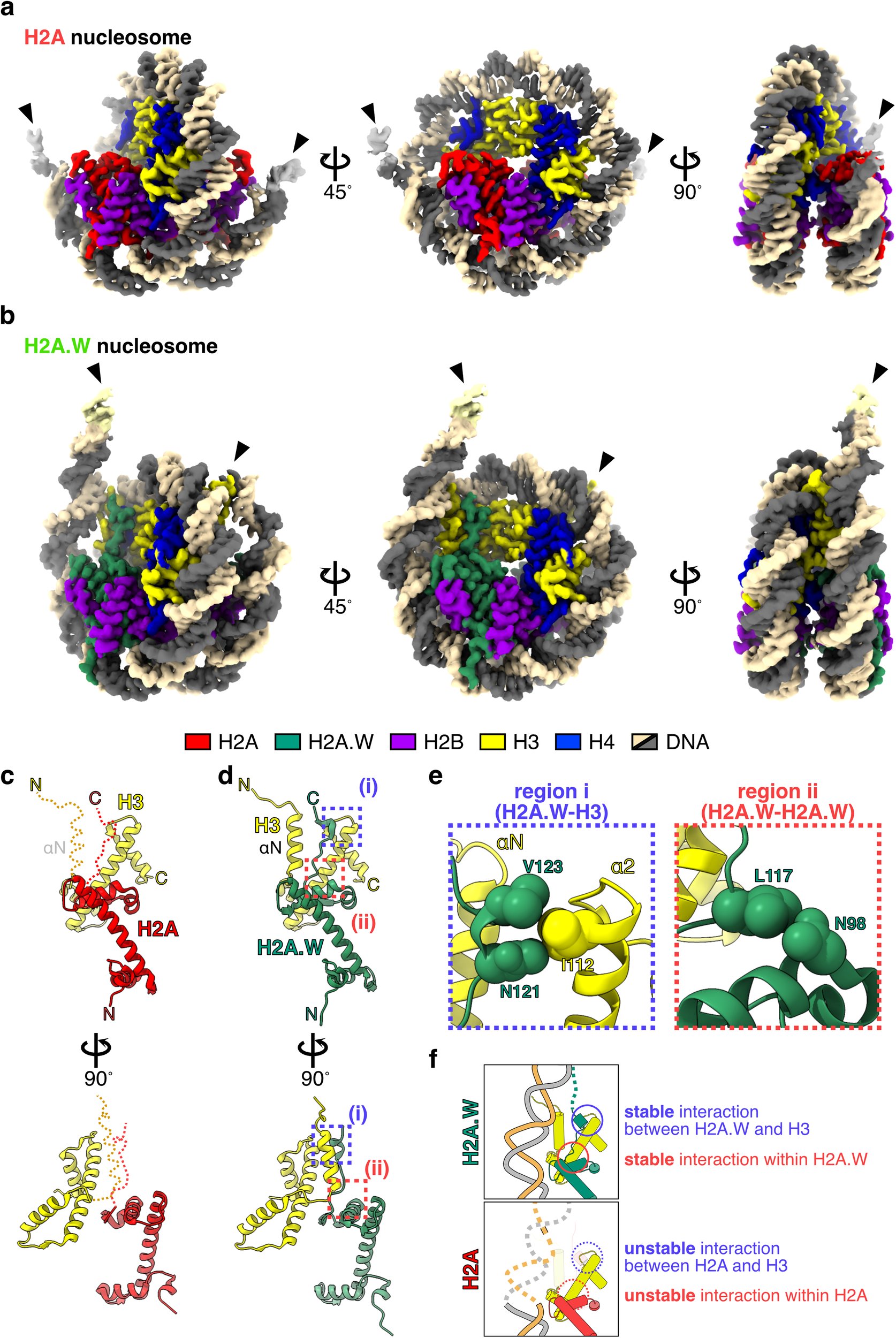
Check out our latest research on chromatin remodeler DDM1 and the H2A.W nucleosome. We were able to visualize how DDM1 opens the the H2A.W nucleosome! This is a great collaboration with Drs Akihisa Osakabe and Tetsuji Kakutani. Congratulations! nature.com/articles/s4146…
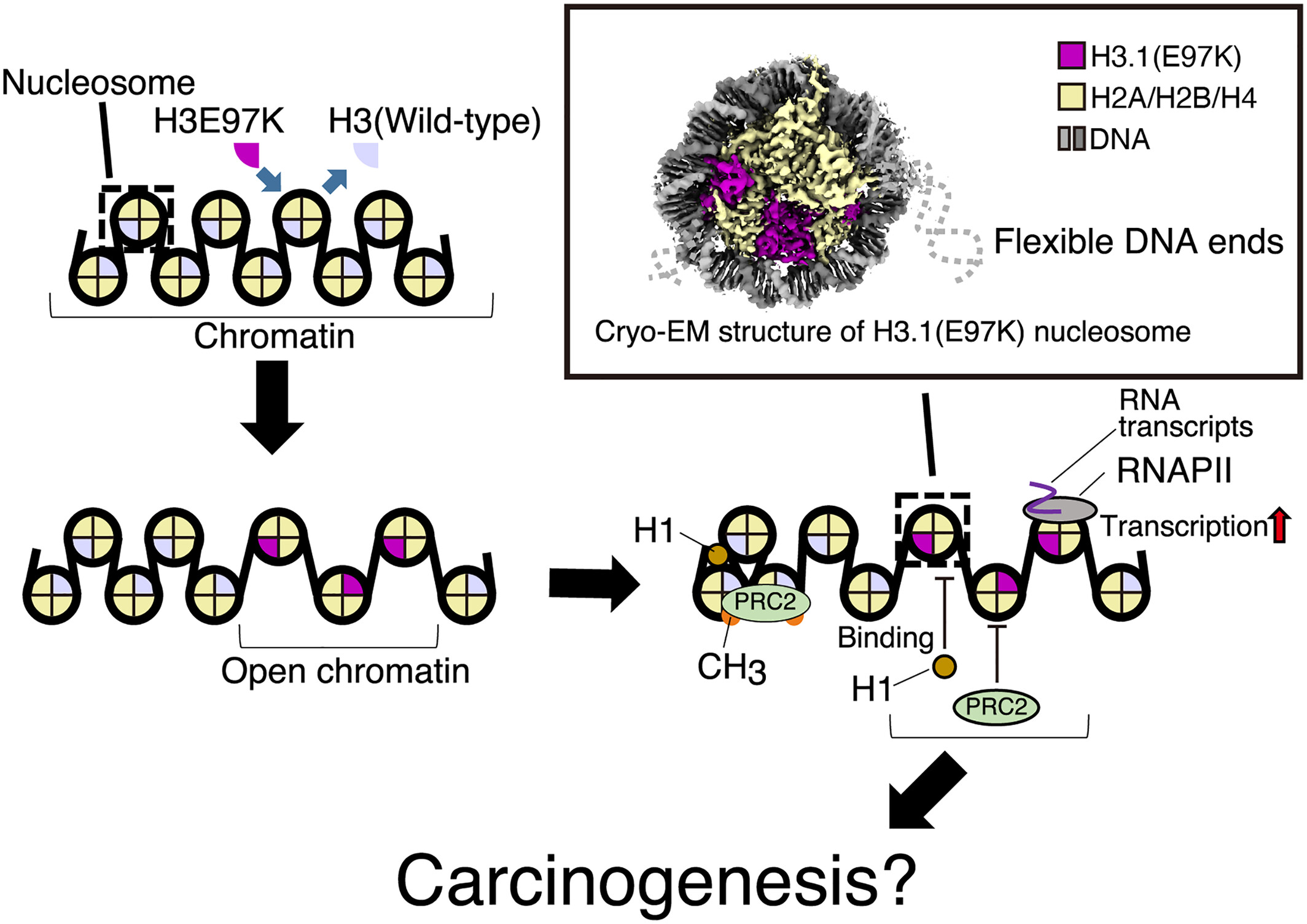
Our new paper about nucleosome containing H3E97K mutation is published! This mutation is found in cancer cells. onlinelibrary.wiley.com/doi/10.1111/gt…