Genome Biology
@GenomeBiology
Tweets about latest articles in the journal Genome Biology, conferences & more. Part of @SpringerNature. Also read our blogs (http://tiny.cc/GBblogs).
🚨 Job alert! If you have expertise in bioinformatics—especially in functional genomics, metagenomics, or epigenetics: join our editorial team! 📍New York, Philly, or Shanghai 🗓️ Apply by Aug 3 🔗lnkd.in/erkUvyib Please share! #Genomics #Bioinformatics #EditorJobs
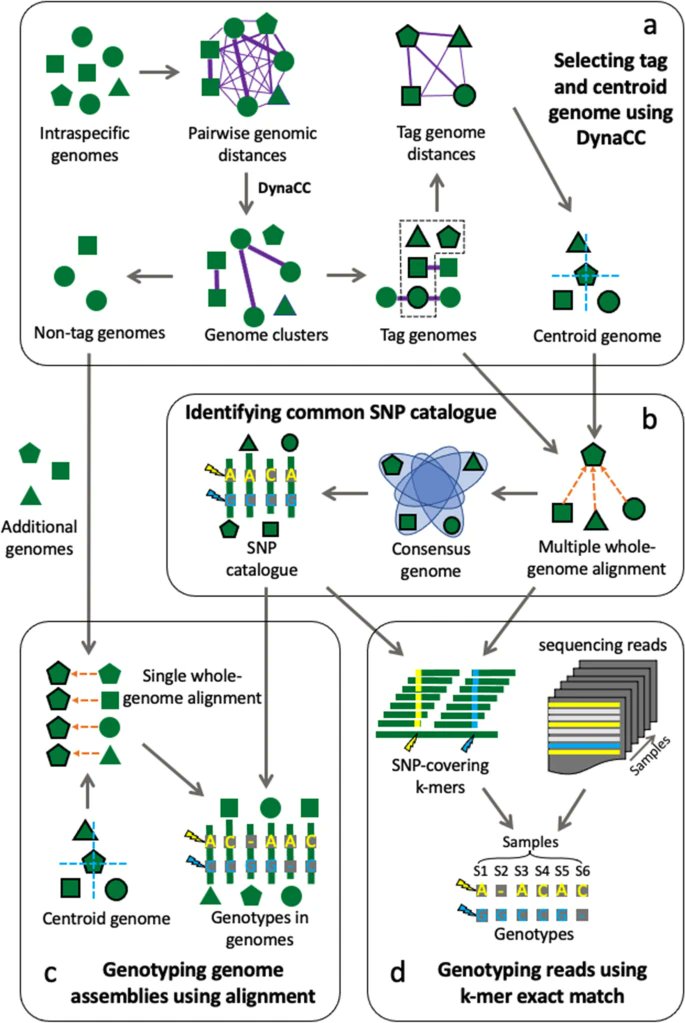
Maast, a bioinformatics tool, presented by Zhou Jason Shi, Stephen Nayfach & Katherine Pollard @czbiohub @jgi implements a novel algorithm to heuristically identify a minimal set of diverse conspecific genomes, enabling rapid and accurate genotyping bit.ly/3s8vw5O
Genome Biology is calling for submissions to the Collection "Genome editing and cancer", guest edited by Emanuel Gonçalves (@emanuelvgo) and Xueqiu Lin (@xueqiu_lin). For more details: biomedcentral.com/collections/ge… #CRISPR #geneediting
📢Join us as an Associate or Senior Editor in either our 🇺🇸NY, Philly or 🇨🇳Shanghai offices. If you have expertise in #singlecell or #spatial #bioinformatics, #crispr, #microbiome or #metagenomics methods -> apply before 22May! 🗓️ more info: performancemanager5.successfactors.eu/sf/jobreq?jobI…
ChIPr, from Abbas, Zhang, @Ram_Mani_PhD & co, a new method to predict 3D chromatin structure from ChIP-seq data. They demonstrate using RAD21 data, and show the method's predictions correlate well with HiC and ChIA-PET results. genomebiology.biomedcentral.com/articles/10.11…
SQANTI-SIM, presented by @anaconesa @ConesaLab & co. enables the simulation of realistic novel transcripts based on the reference annotation, and is an essential tool for the benchmarking of lrRNA-seq methods for new transcript detection bit.ly/3RkPzGZ

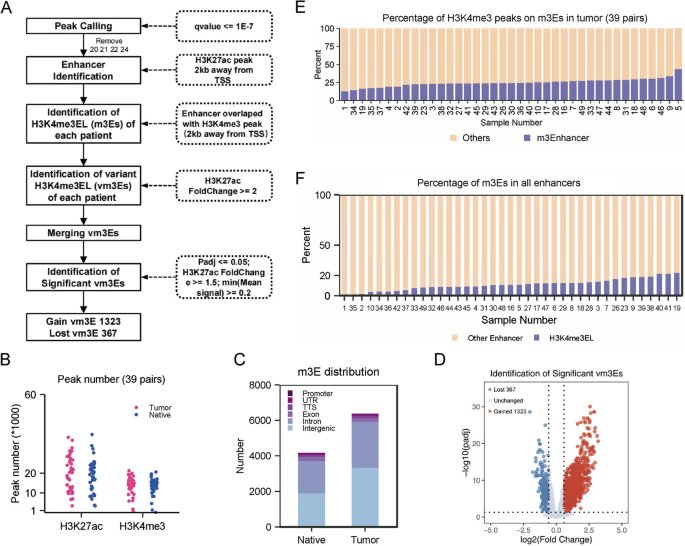
New research from Min Wu, Yong Xiao, Ming-Kai Chen & co. @WHU_1893 illustrating the genome-wide landscape & regulatory mechanisms of m3Es in colorectal cancer, and revealing potential novel strategies for #cancertreatment bit.ly/47P8259
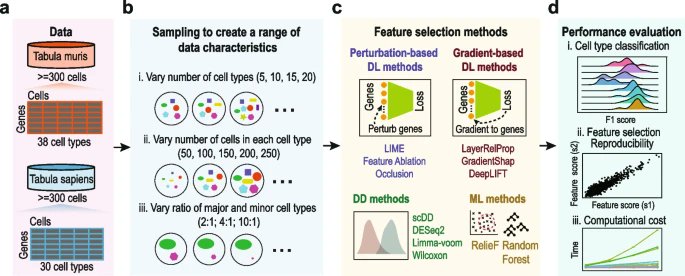
New research from @PengyiYang82 @haohuang1999 & co. @Sydney_Uni providing a reference for future development and application of #deeplearning based feature selection methods for single-cell omics data analyses bit.ly/47xz54N
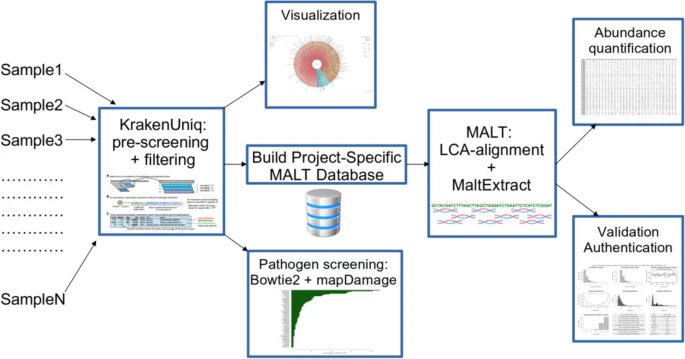
aMeta, presented by @ZoePochon & co. is an accurate metagenomic profiling workflow for ancient DNA designed to minimize the amount of false discoveries and computer memory requirements, likely to be of broad utility for the ancient metagenomics field bit.ly/3QdVblW
DISCERN, presented by @stefan_bonn & co. is a novel deep generative network that precisely reconstructs missing single-cell gene expression using a reference dataset, which can easily be applied to a variety of data sets yielding novel insights bit.ly/3td1kqG
New #cancerresearch from Qian-Fei Wang, Shaoyan Hu & co. revealing leukemia stem cells and OXPHOS as two major chemoresistant features in human AML patients, suggesting important implications for targeting residual chemo-surviving AML cells bit.ly/3qUppSb

GTM-decon, presented by Lakshmipuram Seshadri Swapna, Michael Huang & Yue Li @mcgillu is an efficient marker-free deconvolution method that takes the full advantage of the single-cell RNA-seq reference data for cell-type deconvolution bit.ly/45F0v82
BamQuery, a proteogenomic tool presented by @MariaVPetite @EhxGregory & co. @IRIC_umontreal enables for the first time a uniformization of proteogenomic analyses in MHC-I immunopeptidomics, due to its exhaustivity, speed, ease of use & versatility bit.ly/3OD34QY
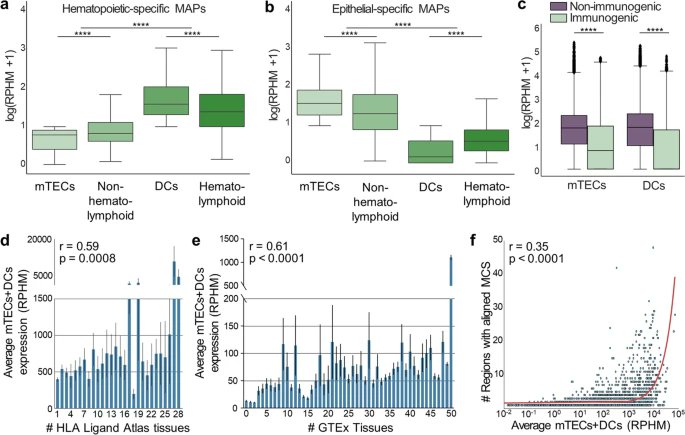
New research from @sergio_vim @jordanatbell & co. @KingsCollegeLon, with findings that improve the characterisation of the mechanisms underlying DNA methylation variability & are informative for prioritisation of GWAS variants for functional follow-ups bit.ly/3qh5pJi
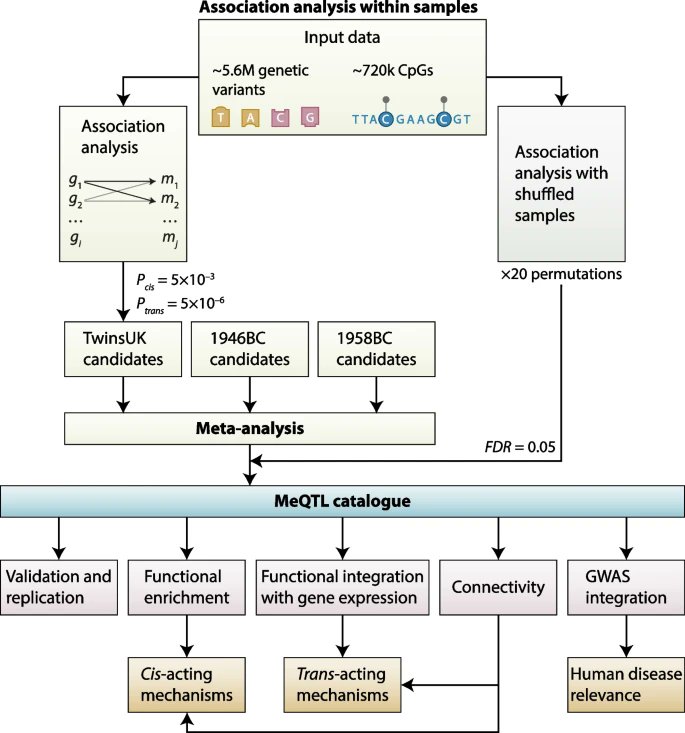
📣 Check out the first published papers @BMC_GPH ! Congrats! Our new sibling is growing up! 🎉😉
We are excited to share that @BMC_GPH has published its first articles. Please explore our first content and share the news …cglobalpublichealth.biomedcentral.com/articles
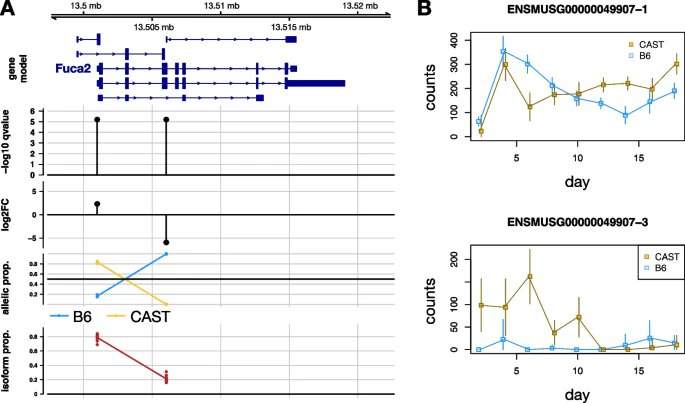
SEESAW, presented by @FennecPaaaw @mikelove & co. @UNC is a new suite of methods for quantifying and testing AI, offering analysis at various levels of resolution compared to existing methods for detecting when there is isoform-level imbalance bit.ly/44SfbQB
Today we launched a collection with the AVE Alliance @varianteffects related to Multiplexed Assays of Variant Effect. This collection also features their vision for a comprehensive atlas characterising the function of every variant in the human genome! biomedcentral.com/collections/to…

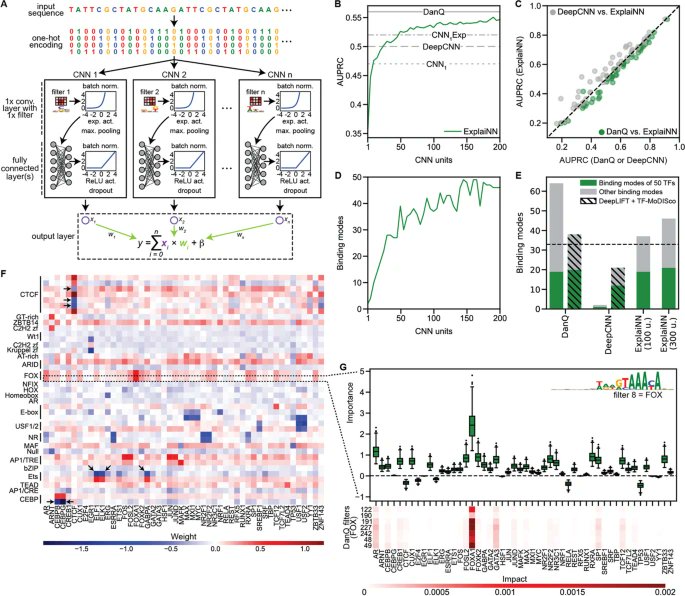
ExplaiNN, presented by @WyWyWa @NovakovskyG @ofornes @manusaraswat10 @sara_mostafavi is a #deeplearning model using an interpretable neural network design, providing both global and local biological insights without compromising on performance bit.ly/46AVheS
New #stemcell research from @RushworthLab & co. @uniofeastanglia @EarlhamInst giving a unique insight into hematopoietic stem cell reactivation upon platelet depletion and of clonal dynamics in both steady state & under stress bit.ly/3CPjfVL

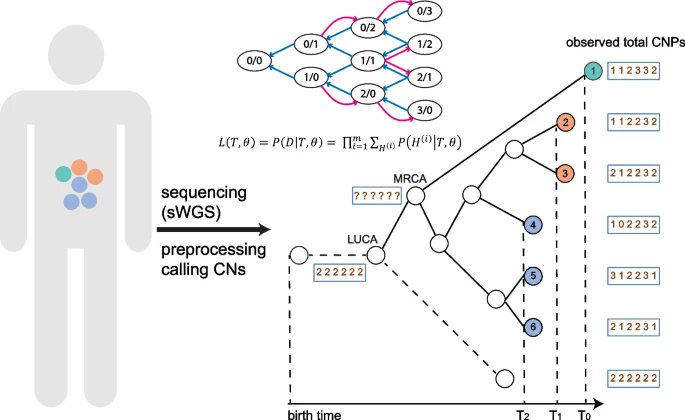
CNETML presented by @lubingxin @yosoykit @trevoragraham @zihengyang @cssb_lab &co @ucl is a new maximum likelihood method to infer phylogenies based on copy number profiles from multiple samples of patients which are helpful to understand #cancerevolution bit.ly/3pbeWBd