Omar S.M. El Nahhas
@ElNahhasOSM
Computer Science PhD Candidate @ Kather Lab for Clinical AI. Researching better methods for cancer biomarker prediction from pathology slides using AI.
Very happy to share our recent article in Nature Cancer on AI Agents for Decision Making in Cancer :) @jnkath
A paper in @NatureCancer presents an autonomous artificial intelligence agent system for deployment of specialized medical oncology computational tools. The AI agent reached correct clinical conclusions in 91% of cases. go.nature.com/4dZKaQC
Featured today by @MSKLibrary: Multimodal histopathologic models stratify hormone receptor-positive early breast cancer #BreastCancer #Histopathology #HormoneReceptorPositive #CancerResearch #PrecisionMedicine @kevinmboehm @ElNahhasOSM @antoniomarraMD @LiorBraunstein…
Good to see this in print. We built Orpheus, a model to infer OncoType score from H&E and pathology text reports, finding that multimodality strengthens inference. We also tuned directly on metastatic recurrence to refine TAILORx low/intermed. risk groups. nature.com/articles/s4146…
Had a great time at #MICCAI2024 🇲🇦, giving an oral presentation in front of a large audience, doing a poster session with dozens of interested people, and connecting with amazing people in the field of medical imaging 🤩 See you at the next edition! 🇰🇷
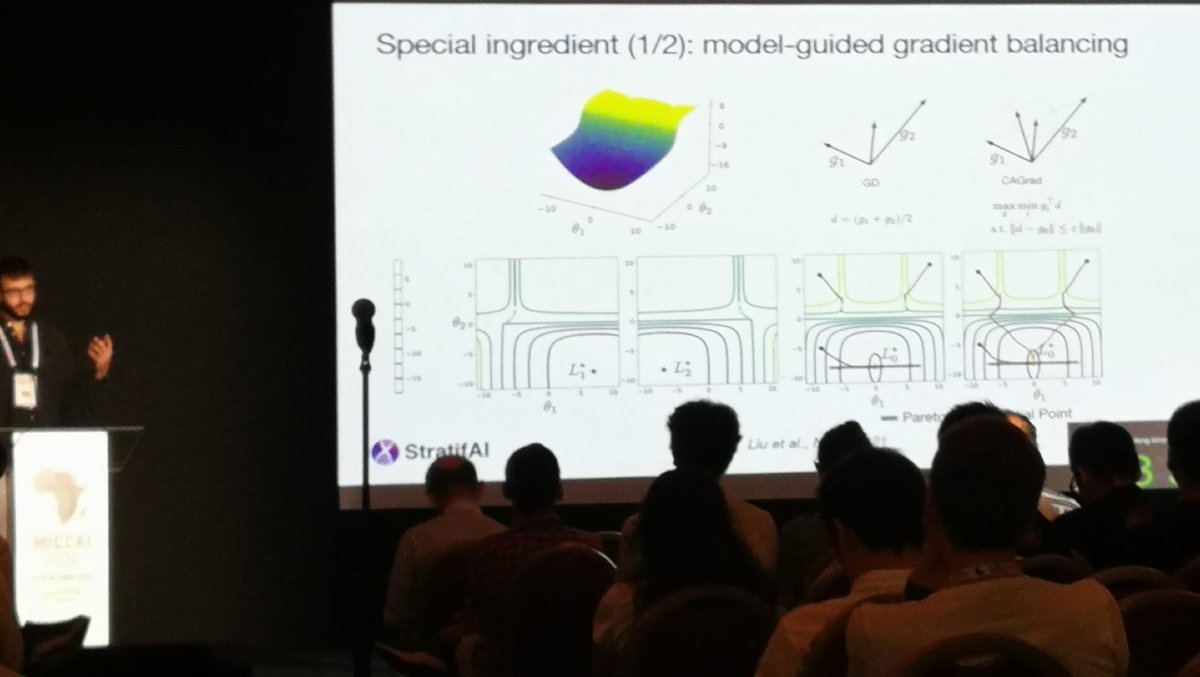
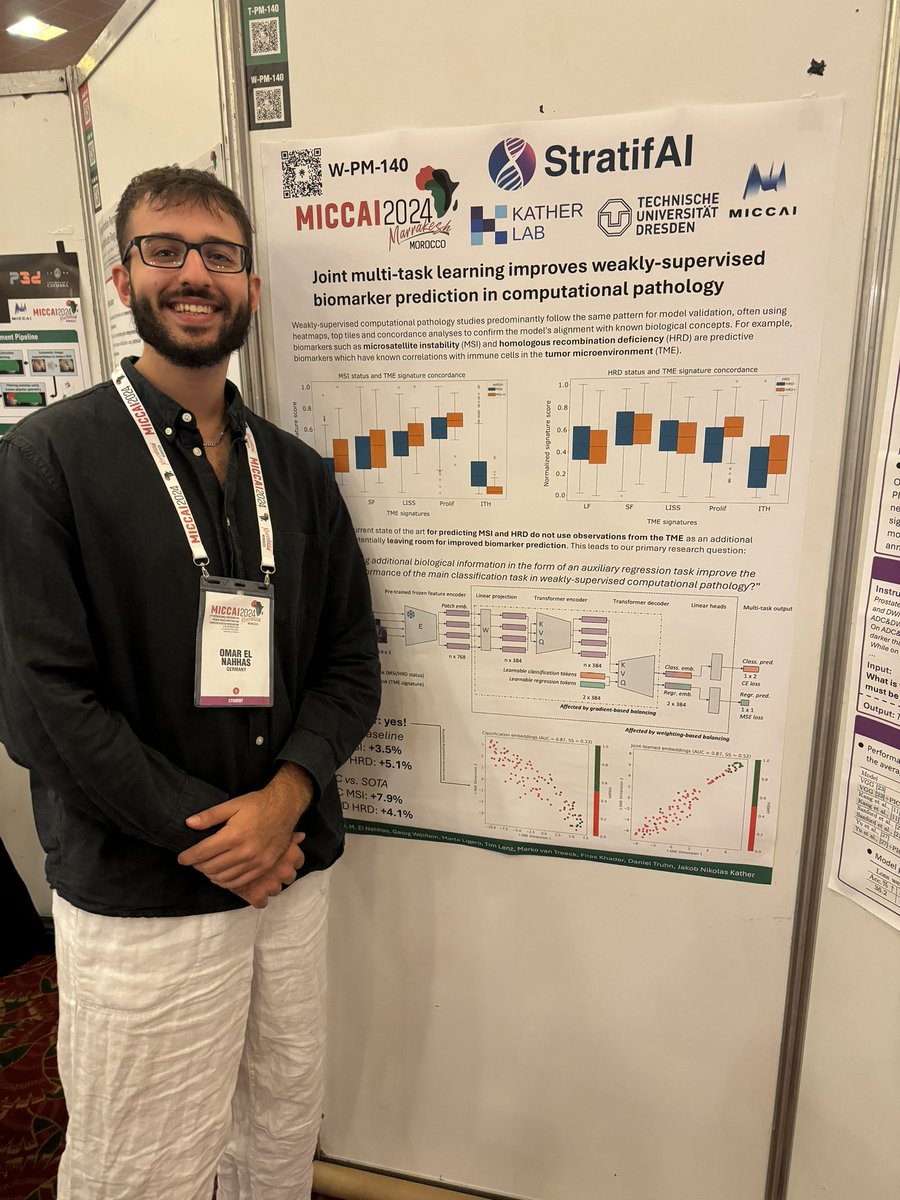


Just arrived at #MICCAI2024 in Marrakesh, and already enjoying the beautiful weather and amazing food 🇲🇦 has to offer. Looking forward to giving my oral presentation on Wednesday, and exchange ideas + get inspired from all the great research work here 🌞


Happy to see our latest @NatureProtocols protocol being included in 'Cancer at Nature Portfolio' - a curated collection of cutting-edge cancer research, methods, and reviews from across Nature journals. Check it out here 👇 nature.com/collections/bp…

New paper from @katherlab led by @ElNahhasOSM et al. 😊👇
#NewNProt moves from whole-slide image to #biomarker prediction: end-to-end weakly-supervised #deeplearning in #computationalpathology bit.ly/3zoUh1w
#NewNProt moves from whole-slide image to #biomarker prediction: end-to-end weakly-supervised #deeplearning in #computationalpathology bit.ly/3zoUh1w
Latest publication out :) Very interesting experience to deploy a software solution on so many hospital systems across the world, which shaped the protocol until what it’s finally become. Thank you to all the collaborators who tested and improved the protocol!🤩
Finally out in @NatureProtocols : our group's workflows for end to end computational pathology. Compatible with UNI and other foundation models. Led by @ElNahhasOSM from @katherlab journal link: nature.com/articles/s4159… full text: rdcu.be/dT35l
Honored to have been invited for an oral presentation in addition to my poster presentation at #MICCAI2024 for my latest paper “Joint multi-task learning improves weakly-supervised biomarker prediction in computational pathology” 🤩 See you there! @katherlab @EKFZdigital
🔊 The #MICCAI2024 preliminary main conference program is now online at: 🔗conferences.miccai.org/2024/files/dow… Congratulations to all of the oral presenters! 👏 We look forward to seeing you in October. @MiccaiStudents @WomenInMICCAI @RMiccai #MICCAI #AI #medicalimaging #DeepLearning
Happy to announce that StratifAI has raised €1.5M in pre-seed funding 🚀 Check out the press release below 👇 t.ly/ydrmF And we're also hiring for roles in Berlin 👇 t.ly/y0F-x

This article (arxiv.org/pdf/2408.15823) from @jnkath is the most comprehensive effort to compare comp path foundation models to date. Showing that multimodal FMs like CONCH outperform unimodal FMs on unimodal tasks, and that the diversity of data >> quantity of data.
New research from @katherlab on #computational #pathology: We benchmark pathology foundation models 👇 Several of these are publicly available under permissive licenses. arxiv.org/abs/2408.15823 Some takeaways: 1. The vision-language model CONCH (@AI4Pathology) outperforms…
New research from @katherlab on #computational #pathology: We benchmark pathology foundation models 👇 Several of these are publicly available under permissive licenses. arxiv.org/abs/2408.15823 Some takeaways: 1. The vision-language model CONCH (@AI4Pathology) outperforms…
Today I had my computer science PhD status talk, a formal requirement enabling submission of the thesis! The examination was a success, and I’m now well-equipped with feedback to finalize my PhD thesis 🎉 Big thank you to my mentor/supervisor @jnkath and @MOSAICgroup1! 🤩


Today marks the end of the #TANGERINE summer school 2024, where I had the pleasure to present the final session on commercializing academic research 🗣️

Excited to present my latest work accepted in #MICCAI2024 on joint multi-task learning in computational pathology! We reach SOTA performance on predicting MSI and HRD status by learning auxiliary regression tasks related to the tumor microenvironment 🔬 arxiv.org/abs/2403.03891
Enjoying the opportunity to meet @ElNahhasOSM in person in the AI course organized by @Caldera60373705 in Paris with @jnkath. Talking about STAMP of course.
Being able to see the field >2 years ahead and trying to convince people in the present is not an easy job! 💭🚀
Back in August 2022, a few months before chatGPT came out, I unsucessfully tried to convince medical journals that large language models (LLMs) are interesting ... Very happy that we were now able to provide quantitative evidence for the utility of LLMs in oncology in @NEJM_AI…
Looking forward to presenting at the AI in Pathology workshop for the 3rd year already! See you there 🤩
Back in August 2022, a few months before chatGPT came out, I unsucessfully tried to convince medical journals that large language models (LLMs) are interesting ... Very happy that we were now able to provide quantitative evidence for the utility of LLMs in oncology in @NEJM_AI…
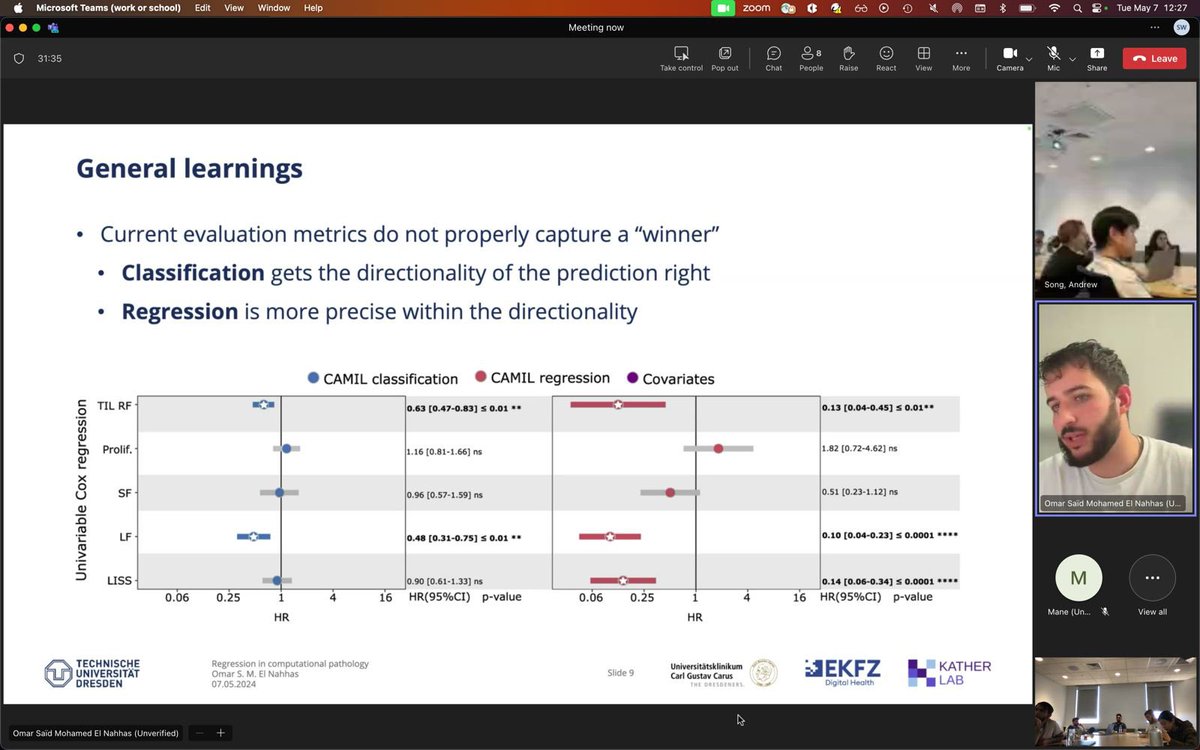
Today I presented and discussed my latest works on regression in computational pathology at the @AI4Pathology lab meeting 🤩. Thank you for the invitation! @sophiajwagner @GreatAndrew90